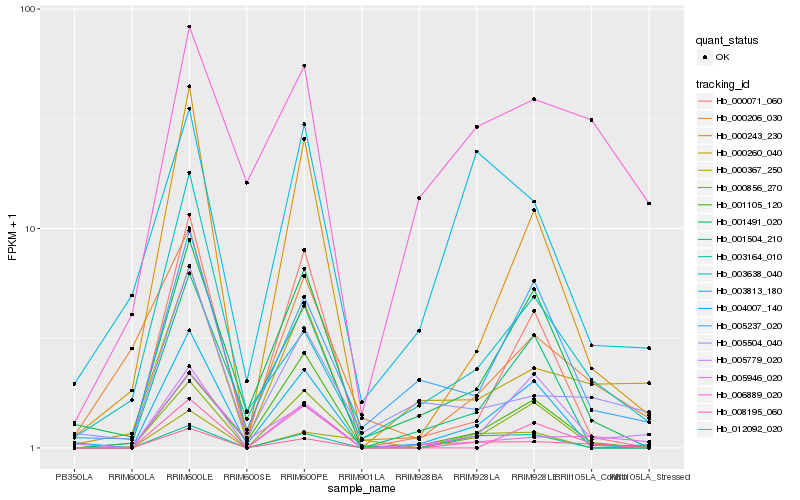
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012092_020 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003164_010 |
0.2359872049 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000243_230 |
0.2531425316 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000856_270 |
0.2581579628 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 5 |
Hb_004007_140 |
0.2742727741 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001491_020 |
0.2784568328 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000071_060 |
0.2838660421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005946_020 |
0.289065482 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |
| 9 |
Hb_003813_180 |
0.2918170109 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 10 |
Hb_000367_250 |
0.2923300764 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_003638_040 |
0.292793757 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_000260_040 |
0.2933963672 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 13 |
Hb_008195_060 |
0.2976904475 |
transcription factor |
TF Family: NAC |
NAC transcription factor 018 [Jatropha curcas] |
| 14 |
Hb_005237_020 |
0.299324984 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 15 |
Hb_001105_120 |
0.2996064456 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001504_210 |
0.3029424137 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006889_020 |
0.3043651609 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 18 |
Hb_005779_020 |
0.3046429644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005504_040 |
0.3083847589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000206_030 |
0.3087901787 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |