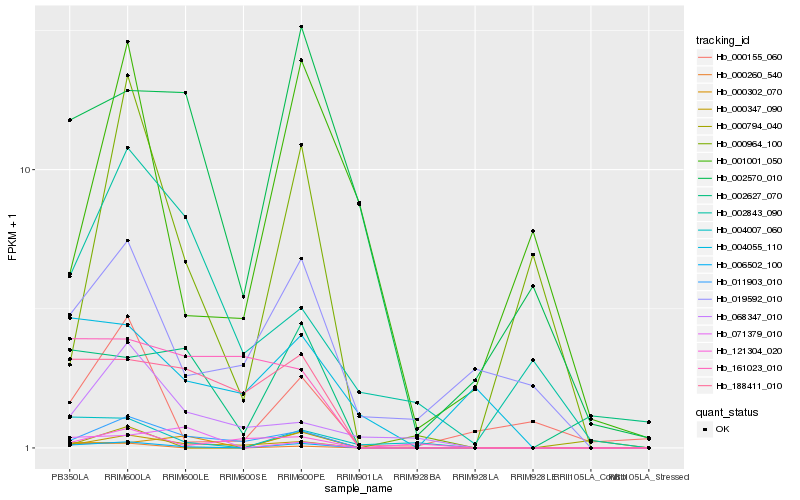
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006502_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s01580g [Populus trichocarpa] |
| 2 |
Hb_011903_010 |
0.2594730298 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_004007_060 |
0.2674545205 |
- |
- |
Ribose-5-phosphate isomerase A [Gossypium arboreum] |
| 4 |
Hb_000347_090 |
0.2681739114 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 5 |
Hb_000260_540 |
0.2785440153 |
- |
- |
hypothetical protein JCGZ_00592 [Jatropha curcas] |
| 6 |
Hb_188411_010 |
0.3193606957 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 7 |
Hb_002627_070 |
0.3338914989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000964_100 |
0.3378543651 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 9 |
Hb_000155_060 |
0.339126523 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_002570_010 |
0.3457616271 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 11 |
Hb_019592_010 |
0.347009129 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 12 |
Hb_000302_070 |
0.3503792666 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_068347_010 |
0.3625351693 |
- |
- |
- |
| 14 |
Hb_004055_110 |
0.3636482382 |
- |
- |
PREDICTED: uncharacterized protein LOC105631442 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000794_040 |
0.3761688501 |
- |
- |
PREDICTED: RING-H2 finger protein ATL29-like [Jatropha curcas] |
| 16 |
Hb_071379_010 |
0.3827220881 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 3-like [Nelumbo nucifera] |
| 17 |
Hb_002843_090 |
0.3835973711 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_161023_010 |
0.3852806418 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |
| 19 |
Hb_001001_050 |
0.3864868857 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 20 |
Hb_121304_020 |
0.3876408421 |
- |
- |
- |