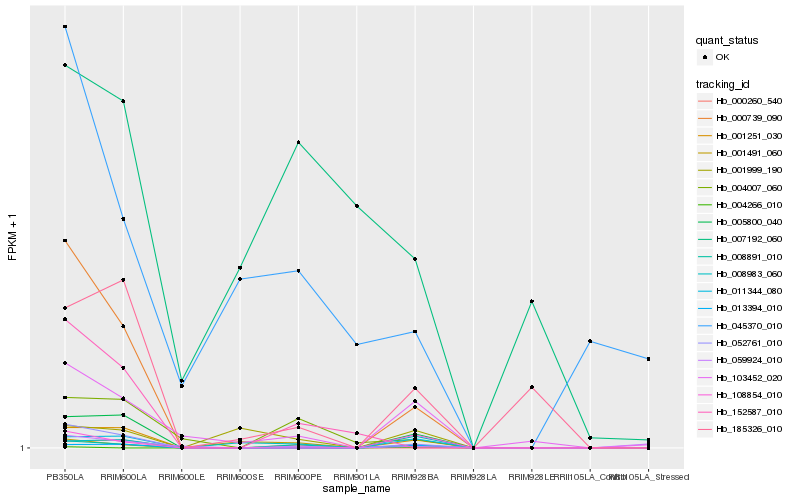
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008891_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_D01145 [Eucalyptus grandis] |
| 2 |
Hb_004007_060 |
0.2746466544 |
- |
- |
Ribose-5-phosphate isomerase A [Gossypium arboreum] |
| 3 |
Hb_103452_020 |
0.2982424595 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_000260_540 |
0.3309410089 |
- |
- |
hypothetical protein JCGZ_00592 [Jatropha curcas] |
| 5 |
Hb_001251_030 |
0.3405411018 |
- |
- |
hypothetical protein PHAVU_002G189800g [Phaseolus vulgaris] |
| 6 |
Hb_005800_040 |
0.3429764931 |
- |
- |
- |
| 7 |
Hb_052761_010 |
0.3437748875 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 8 |
Hb_013394_010 |
0.3473666387 |
- |
- |
PREDICTED: laccase-14-like [Malus domestica] |
| 9 |
Hb_011344_080 |
0.3493233247 |
- |
- |
- |
| 10 |
Hb_000739_090 |
0.3515503722 |
- |
- |
- |
| 11 |
Hb_108854_010 |
0.3567077593 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 12 |
Hb_008983_060 |
0.3635655631 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance protein, putative [Theobroma cacao] |
| 13 |
Hb_152587_010 |
0.364538108 |
- |
- |
- |
| 14 |
Hb_185326_010 |
0.3671081547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004266_010 |
0.3677129812 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_001999_190 |
0.3768790456 |
- |
- |
GDP-mannose transporter, putative [Ricinus communis] |
| 17 |
Hb_001491_060 |
0.3789401563 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6-like isoform X1 [Populus euphratica] |
| 18 |
Hb_059924_010 |
0.3811209769 |
- |
- |
- |
| 19 |
Hb_007192_060 |
0.3888999091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_045370_010 |
0.4009950391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |