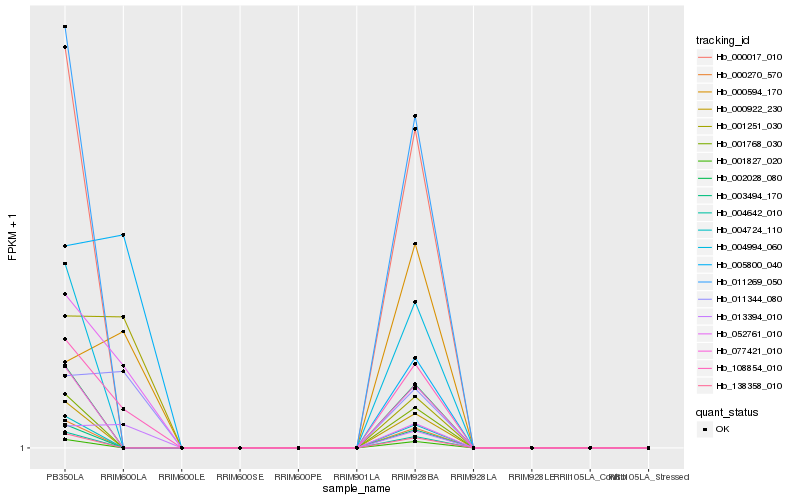
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108854_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 2 |
Hb_011344_080 |
0.1986719747 |
- |
- |
- |
| 3 |
Hb_013394_010 |
0.2047091396 |
- |
- |
PREDICTED: laccase-14-like [Malus domestica] |
| 4 |
Hb_000594_170 |
0.245088963 |
- |
- |
- |
| 5 |
Hb_005800_040 |
0.2653568844 |
- |
- |
- |
| 6 |
Hb_001251_030 |
0.26770381 |
- |
- |
hypothetical protein PHAVU_002G189800g [Phaseolus vulgaris] |
| 7 |
Hb_052761_010 |
0.2911993456 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 8 |
Hb_077421_010 |
0.2958266542 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 9 |
Hb_002028_080 |
0.2958271536 |
- |
- |
hypothetical protein JCGZ_07571 [Jatropha curcas] |
| 10 |
Hb_011269_050 |
0.2958284153 |
- |
- |
- |
| 11 |
Hb_000017_010 |
0.295828458 |
- |
- |
- |
| 12 |
Hb_003494_170 |
0.2958300707 |
- |
- |
- |
| 13 |
Hb_001827_020 |
0.2958323458 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_004994_060 |
0.2958530028 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 15 |
Hb_001768_030 |
0.2958809178 |
- |
- |
- |
| 16 |
Hb_000922_230 |
0.2959182906 |
- |
- |
- |
| 17 |
Hb_004724_110 |
0.2960157032 |
- |
- |
alpha-1,2-fucosyltransferase [Populus tremula x Populus alba] |
| 18 |
Hb_000270_570 |
0.2960512118 |
- |
- |
PREDICTED: peroxidase 65-like [Jatropha curcas] |
| 19 |
Hb_004642_010 |
0.2961558319 |
- |
- |
Integrase core domain, putative [Oryza sativa Japonica Group] |
| 20 |
Hb_138358_010 |
0.2961723331 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |