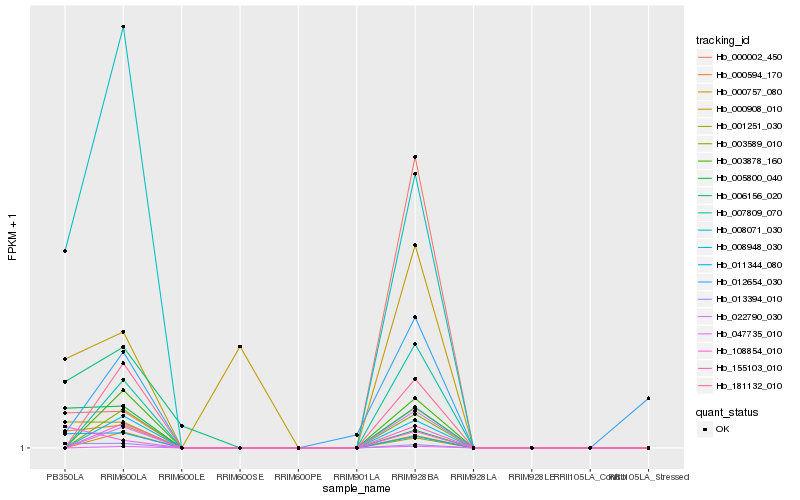
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_170 |
0.0 |
- |
- |
- |
| 2 |
Hb_011344_080 |
0.203688157 |
- |
- |
- |
| 3 |
Hb_013394_010 |
0.2153531904 |
- |
- |
PREDICTED: laccase-14-like [Malus domestica] |
| 4 |
Hb_108854_010 |
0.245088963 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 5 |
Hb_008071_030 |
0.2632083506 |
- |
- |
- |
| 6 |
Hb_000908_010 |
0.3128539066 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_005800_040 |
0.3132790704 |
- |
- |
- |
| 8 |
Hb_001251_030 |
0.3283521695 |
- |
- |
hypothetical protein PHAVU_002G189800g [Phaseolus vulgaris] |
| 9 |
Hb_022790_030 |
0.333291836 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 10 |
Hb_007809_070 |
0.3338291335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_047735_010 |
0.3454438824 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 12 |
Hb_000002_450 |
0.3454686148 |
- |
- |
hypothetical protein JCGZ_02285 [Jatropha curcas] |
| 13 |
Hb_155103_010 |
0.362303724 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_006156_020 |
0.3626972265 |
- |
- |
- |
| 15 |
Hb_003589_010 |
0.3635409966 |
- |
- |
- |
| 16 |
Hb_008948_030 |
0.3643527932 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 17 |
Hb_003878_160 |
0.3658866524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_012654_030 |
0.3701622379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_181132_010 |
0.3716531647 |
- |
- |
- |
| 20 |
Hb_000757_080 |
0.3731003804 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |