| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
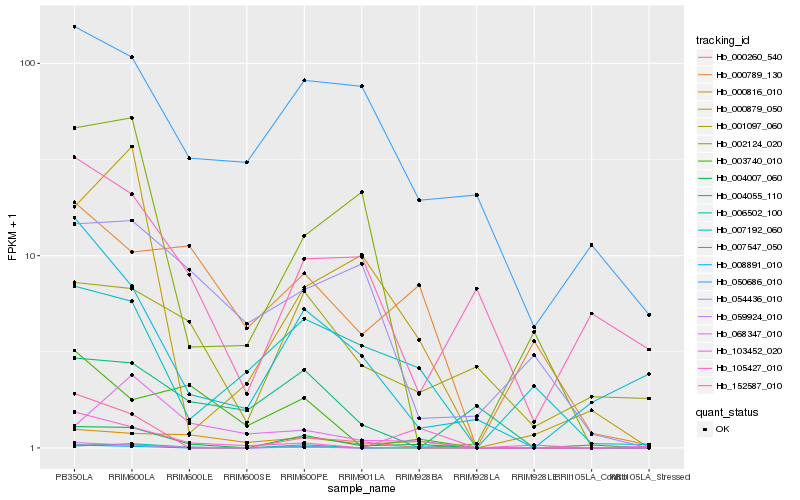
Hb_004007_060 |
0.0 |
- |
- |
Ribose-5-phosphate isomerase A [Gossypium arboreum] |
| 2 |
Hb_006502_100 |
0.2674545205 |
- |
- |
hypothetical protein POPTR_0003s01580g [Populus trichocarpa] |
| 3 |
Hb_008891_010 |
0.2746466544 |
- |
- |
hypothetical protein EUGRSUZ_D01145 [Eucalyptus grandis] |
| 4 |
Hb_000260_540 |
0.2791994389 |
- |
- |
hypothetical protein JCGZ_00592 [Jatropha curcas] |
| 5 |
Hb_000816_010 |
0.2869830255 |
- |
- |
PREDICTED: uncharacterized protein At2g40430 isoform X2 [Jatropha curcas] |
| 6 |
Hb_152587_010 |
0.3017530508 |
- |
- |
- |
| 7 |
Hb_000879_050 |
0.3060332946 |
- |
- |
PREDICTED: uncharacterized protein LOC105107600 [Populus euphratica] |
| 8 |
Hb_007547_050 |
0.307413235 |
- |
- |
Auxin response factor 9 [Aegilops tauschii] |
| 9 |
Hb_007192_060 |
0.3139360599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002124_020 |
0.3162760631 |
transcription factor |
TF Family: bHLH |
lMYC3 [Hevea brasiliensis] |
| 11 |
Hb_001097_060 |
0.3243999395 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 12 |
Hb_050686_010 |
0.3248285707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000789_130 |
0.3291308718 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 14 |
Hb_003740_010 |
0.3319884303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_105427_010 |
0.3435254729 |
- |
- |
- |
| 16 |
Hb_068347_010 |
0.3488638604 |
- |
- |
- |
| 17 |
Hb_059924_010 |
0.3488997341 |
- |
- |
- |
| 18 |
Hb_004055_110 |
0.349223364 |
- |
- |
PREDICTED: uncharacterized protein LOC105631442 isoform X1 [Jatropha curcas] |
| 19 |
Hb_103452_020 |
0.3527985075 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 20 |
Hb_054436_010 |
0.3571182619 |
- |
- |
PREDICTED: uncharacterized protein LOC105638666 [Jatropha curcas] |