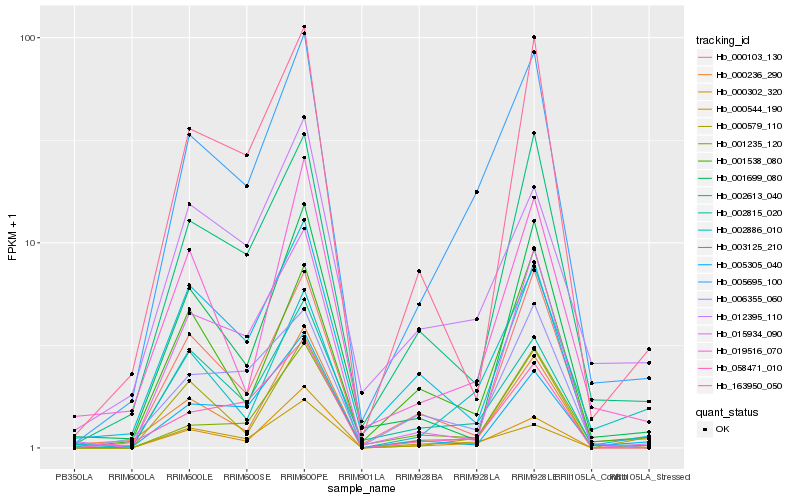
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005695_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638839 [Jatropha curcas] |
| 2 |
Hb_002613_040 |
0.140767577 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 3 |
Hb_002815_020 |
0.1434576062 |
transcription factor |
TF Family: G2-like |
Homeodomain-like superfamily protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_000103_130 |
0.1536477029 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 5 |
Hb_000544_190 |
0.1566736119 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 6 |
Hb_058471_010 |
0.1589036471 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 7 |
Hb_006355_060 |
0.1594600831 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000236_290 |
0.1638053505 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 9 |
Hb_000302_320 |
0.1658028519 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 10 |
Hb_001235_120 |
0.1663263404 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 11 |
Hb_001538_080 |
0.1687969827 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_163950_050 |
0.1691082774 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_012395_110 |
0.1743039051 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 14 |
Hb_000579_110 |
0.1756159148 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 15 |
Hb_019516_070 |
0.1778179707 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 16 |
Hb_002886_010 |
0.1781918999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001699_080 |
0.1782370268 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_005305_040 |
0.1786587867 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_003125_210 |
0.1788417867 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 20 |
Hb_015934_090 |
0.1788696333 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |