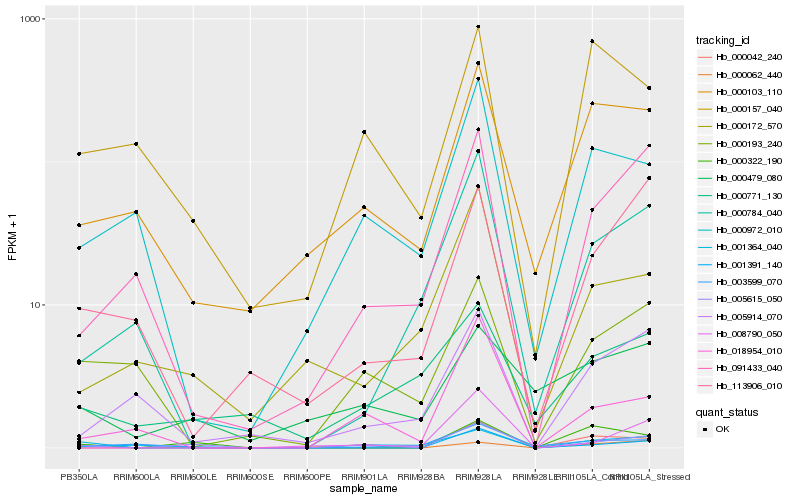
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005615_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636967 [Jatropha curcas] |
| 2 |
Hb_001391_140 |
0.267963492 |
- |
- |
PREDICTED: uncharacterized protein LOC101293660 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_018954_010 |
0.2761371056 |
- |
- |
- |
| 4 |
Hb_001364_040 |
0.2832636792 |
- |
- |
- |
| 5 |
Hb_000322_190 |
0.2852752268 |
- |
- |
- |
| 6 |
Hb_091433_040 |
0.2876646301 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 7 |
Hb_008790_050 |
0.2977255202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000172_570 |
0.298358412 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000103_110 |
0.2988116492 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000972_010 |
0.3080372266 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 11 |
Hb_000062_440 |
0.3136861387 |
- |
- |
hypothetical protein JCGZ_17832 [Jatropha curcas] |
| 12 |
Hb_000193_240 |
0.314873026 |
- |
- |
hypothetical protein CISIN_1g0449881mg, partial [Citrus sinensis] |
| 13 |
Hb_000784_040 |
0.3149559969 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 14 |
Hb_000771_130 |
0.3183949349 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_005914_070 |
0.3212447248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000479_080 |
0.3223848191 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_003599_070 |
0.3228178179 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000042_240 |
0.3237965948 |
- |
- |
- |
| 19 |
Hb_113906_010 |
0.3247499091 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 20 |
Hb_000157_040 |
0.326104211 |
- |
- |
PREDICTED: cycloartenol-C-24-methyltransferase [Jatropha curcas] |