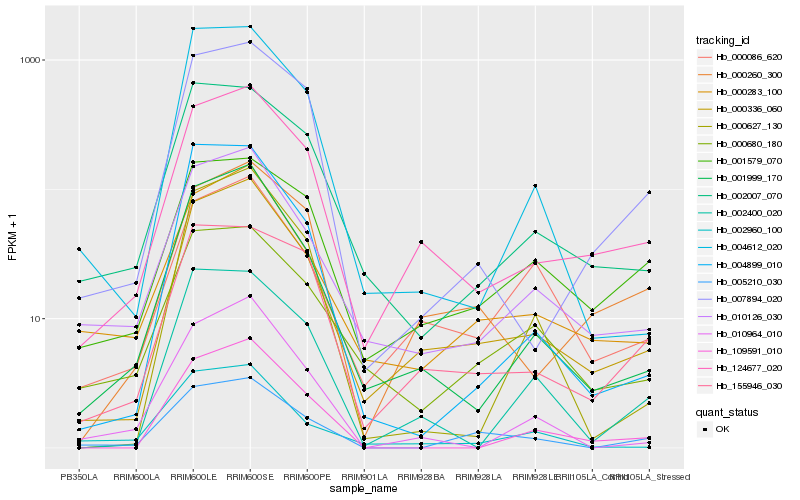
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005210_030 |
0.0 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 2 |
Hb_000336_060 |
0.1673008821 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 3 |
Hb_000283_100 |
0.177122569 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000680_180 |
0.1820938115 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 5 |
Hb_010126_030 |
0.1831752603 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002007_070 |
0.1895305832 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 7 |
Hb_001579_070 |
0.1932616222 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 8 |
Hb_155946_030 |
0.193417899 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 9 |
Hb_007894_020 |
0.1956636836 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 10 |
Hb_001999_170 |
0.2001835137 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 11 |
Hb_124677_020 |
0.201604894 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 12 |
Hb_004899_010 |
0.2033860239 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 13 |
Hb_004612_020 |
0.2046161062 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 14 |
Hb_002960_100 |
0.2080836194 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 15 |
Hb_000260_300 |
0.2111529128 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 16 |
Hb_000086_620 |
0.2112308138 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000627_130 |
0.2131205288 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 18 |
Hb_109591_010 |
0.2132183344 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_010964_010 |
0.2136798574 |
- |
- |
- |
| 20 |
Hb_002400_020 |
0.2151763045 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |