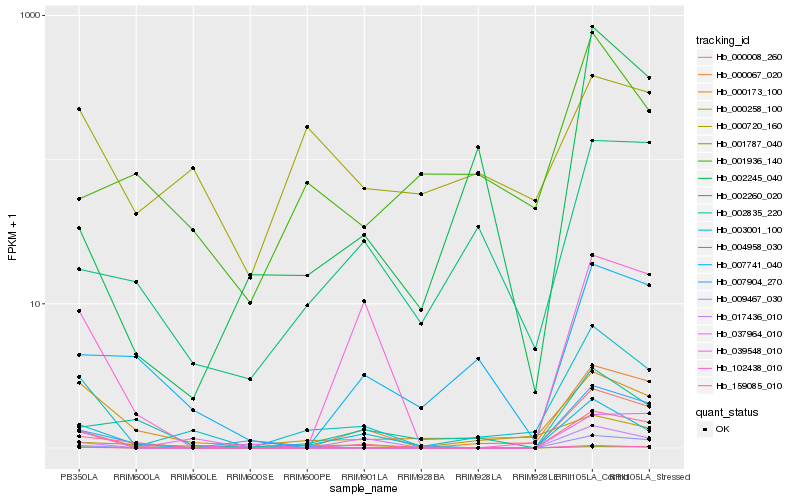
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003001_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103334281 [Prunus mume] |
| 2 |
Hb_000173_100 |
0.2260385709 |
- |
- |
- |
| 3 |
Hb_004958_030 |
0.2550652284 |
- |
- |
PREDICTED: uncharacterized protein LOC104904188 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000067_020 |
0.2619832663 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: inactive disease resistance protein RPS4-like [Jatropha curcas] |
| 5 |
Hb_102438_010 |
0.2703239722 |
- |
- |
- |
| 6 |
Hb_159085_010 |
0.2812563221 |
- |
- |
- |
| 7 |
Hb_000008_260 |
0.2868696754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000258_100 |
0.28850265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009467_030 |
0.2969765733 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 10 |
Hb_000720_160 |
0.3027995156 |
- |
- |
PREDICTED: C2 domain-containing protein At1g63220 [Jatropha curcas] |
| 11 |
Hb_037964_010 |
0.3030059928 |
- |
- |
hypothetical protein MIMGU_mgv1a000906mg [Erythranthe guttata] |
| 12 |
Hb_001787_040 |
0.3040067769 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_039548_010 |
0.3070808333 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 14 |
Hb_007904_270 |
0.3105838099 |
- |
- |
- |
| 15 |
Hb_001936_140 |
0.3112836833 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 1 [Jatropha curcas] |
| 16 |
Hb_007741_040 |
0.3116959954 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105643153 [Jatropha curcas] |
| 17 |
Hb_002835_220 |
0.3119485379 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 18 |
Hb_017436_010 |
0.3137580925 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_002245_040 |
0.3186898149 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 20 |
Hb_002260_020 |
0.3206475556 |
- |
- |
- |