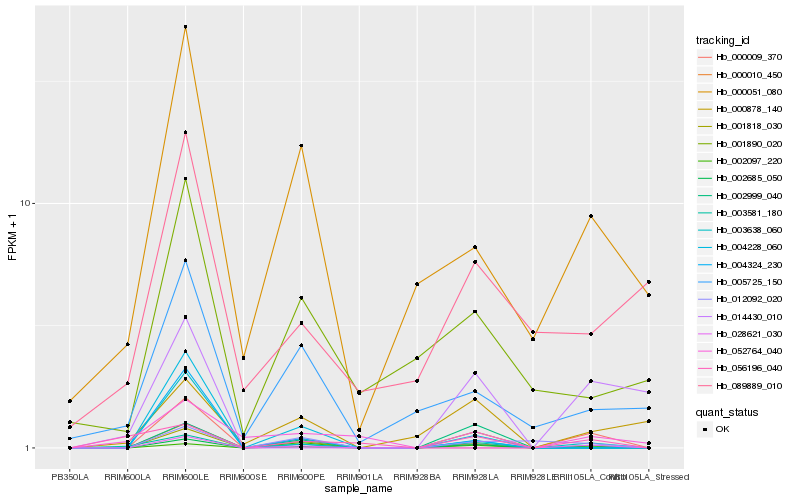
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002685_050 |
0.0 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003638_060 |
0.3023662439 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 3 |
Hb_002097_220 |
0.3084420271 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Sesamum indicum] |
| 4 |
Hb_001818_030 |
0.3097574766 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 5 |
Hb_012092_020 |
0.3194230871 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_002999_040 |
0.320501547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000010_450 |
0.3286317249 |
- |
- |
purine permease, putative [Ricinus communis] |
| 8 |
Hb_003581_180 |
0.3307760513 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 9 |
Hb_000051_080 |
0.3349738731 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 10 |
Hb_000009_370 |
0.3374743559 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 11 |
Hb_028621_030 |
0.3375283149 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 12 |
Hb_004228_060 |
0.3403392392 |
- |
- |
Tubulin beta-2 chain [Triticum urartu] |
| 13 |
Hb_000878_140 |
0.3527468004 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 14 |
Hb_014430_010 |
0.358018645 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_004324_230 |
0.3605105917 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 16 |
Hb_005725_150 |
0.36806196 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_056196_040 |
0.3687527272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001890_020 |
0.3721014774 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 19 |
Hb_052764_040 |
0.3770397674 |
- |
- |
syntaxin 125 family protein [Populus trichocarpa] |
| 20 |
Hb_089889_010 |
0.3864120041 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |