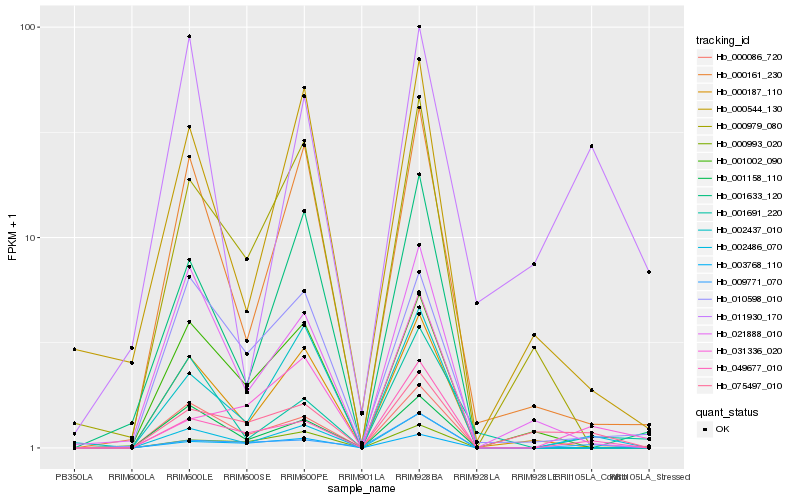
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002486_070 |
0.0 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0008s07280g [Populus trichocarpa] |
| 2 |
Hb_075497_010 |
0.2232107816 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 3 |
Hb_049677_010 |
0.233785242 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_000161_230 |
0.2520162318 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 5 |
Hb_009771_070 |
0.2548057981 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 6 |
Hb_011930_170 |
0.2555181164 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 7 |
Hb_000187_110 |
0.257461503 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 8 |
Hb_010598_010 |
0.2677640319 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 9 |
Hb_001158_110 |
0.2691867699 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 10 |
Hb_000544_130 |
0.2711252895 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 11 |
Hb_002437_010 |
0.2727524566 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 12 |
Hb_001633_120 |
0.2742578066 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 13 |
Hb_000086_720 |
0.2748431619 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_001002_090 |
0.2772145496 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 15 |
Hb_000979_080 |
0.2776803407 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 16 |
Hb_000993_020 |
0.2785660423 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 17 |
Hb_003768_110 |
0.2795435109 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 18 |
Hb_001691_220 |
0.2818310059 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 19 |
Hb_031336_020 |
0.2821474425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_021888_010 |
0.2822144845 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |