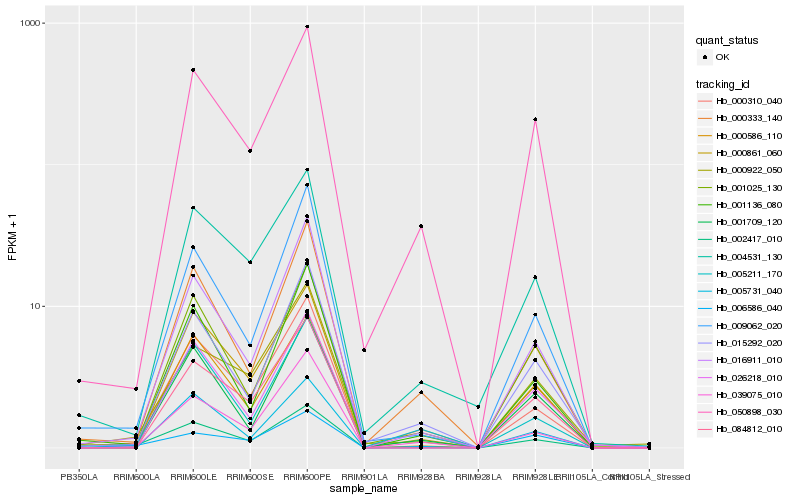
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002417_010 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 2 |
Hb_000310_040 |
0.1137784577 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_016911_010 |
0.119318591 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 4 |
Hb_000861_060 |
0.1219368084 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 5 |
Hb_009062_020 |
0.1303355476 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 6 |
Hb_000586_110 |
0.1346437429 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_039075_010 |
0.1349718496 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_084812_010 |
0.1365094093 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 9 |
Hb_050898_030 |
0.1373299773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001136_080 |
0.1381012205 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 11 |
Hb_001025_130 |
0.1383935628 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 12 |
Hb_000922_050 |
0.1391977355 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 13 |
Hb_001709_120 |
0.1392103948 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 14 |
Hb_015292_020 |
0.1392818503 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_006586_040 |
0.1402460835 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |
| 16 |
Hb_000333_140 |
0.1414883677 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_005731_040 |
0.142595841 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 18 |
Hb_005211_170 |
0.1444458086 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 19 |
Hb_004531_130 |
0.1447732431 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_026218_010 |
0.1456061993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |