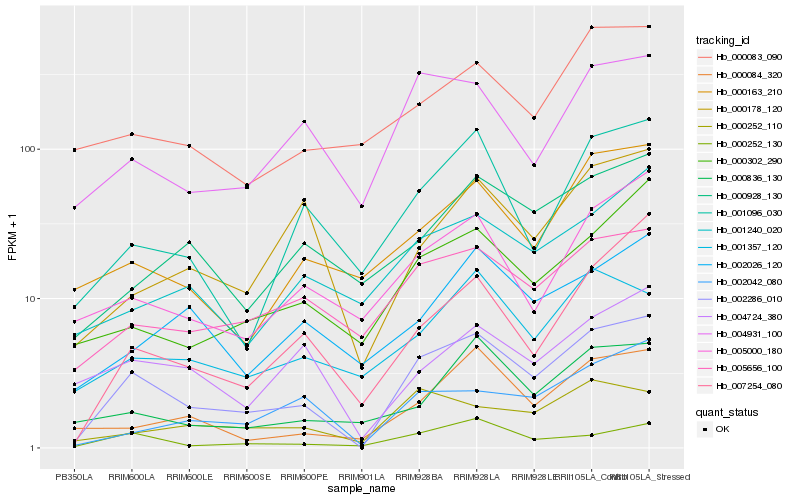
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002286_010 |
0.0 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 2 |
Hb_000252_130 |
0.1806739743 |
- |
- |
hypothetical protein POPTR_0011s17240g, partial [Populus trichocarpa] |
| 3 |
Hb_005656_100 |
0.1828159427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004931_100 |
0.1914405873 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000302_290 |
0.1929591737 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 6 |
Hb_002042_080 |
0.1945807202 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 7 |
Hb_001096_030 |
0.1975168904 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 8 |
Hb_000084_320 |
0.1984227479 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 9 |
Hb_005000_180 |
0.1995124009 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 10 |
Hb_000163_210 |
0.1997351486 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 11 |
Hb_007254_080 |
0.2002465225 |
- |
- |
- |
| 12 |
Hb_000252_110 |
0.2013545481 |
- |
- |
- |
| 13 |
Hb_001240_020 |
0.2019547118 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 14 |
Hb_001357_120 |
0.2031064365 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 15 |
Hb_000928_130 |
0.2033870279 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_004724_380 |
0.2037567223 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 17 |
Hb_000836_130 |
0.2051231889 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000178_120 |
0.2060575825 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 19 |
Hb_002026_120 |
0.2074379143 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 20 |
Hb_000083_090 |
0.2078290064 |
- |
- |
immunophilin, putative [Ricinus communis] |