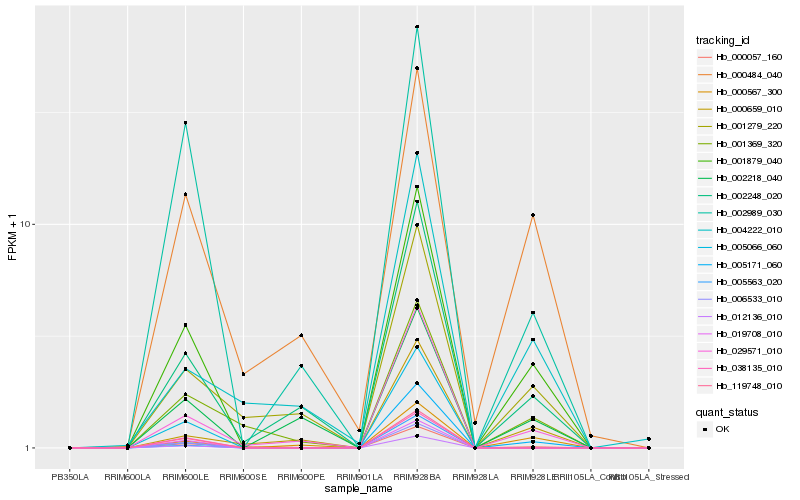
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001879_040 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0033281mg, partial [Citrus sinensis] |
| 2 |
Hb_029571_010 |
0.1351445766 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 3 |
Hb_002248_020 |
0.1551014188 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 [Populus euphratica] |
| 4 |
Hb_002989_030 |
0.1660493186 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 5 |
Hb_000567_300 |
0.1729094034 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 6 |
Hb_001369_320 |
0.1818981868 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 7 |
Hb_001279_220 |
0.1885800434 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_005171_060 |
0.1894990417 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_000484_040 |
0.1911252173 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000057_160 |
0.1944087931 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |
| 11 |
Hb_006533_010 |
0.1997838512 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 12 |
Hb_005066_060 |
0.1999615292 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 13 |
Hb_012136_010 |
0.1999899217 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 14 |
Hb_119748_010 |
0.2012636953 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 15 |
Hb_000659_010 |
0.2024702503 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_019708_010 |
0.2042603809 |
- |
- |
- |
| 17 |
Hb_005563_020 |
0.2043130136 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 18 |
Hb_002218_040 |
0.2043647836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_038135_010 |
0.2051313214 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 20 |
Hb_004222_010 |
0.2076972929 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |