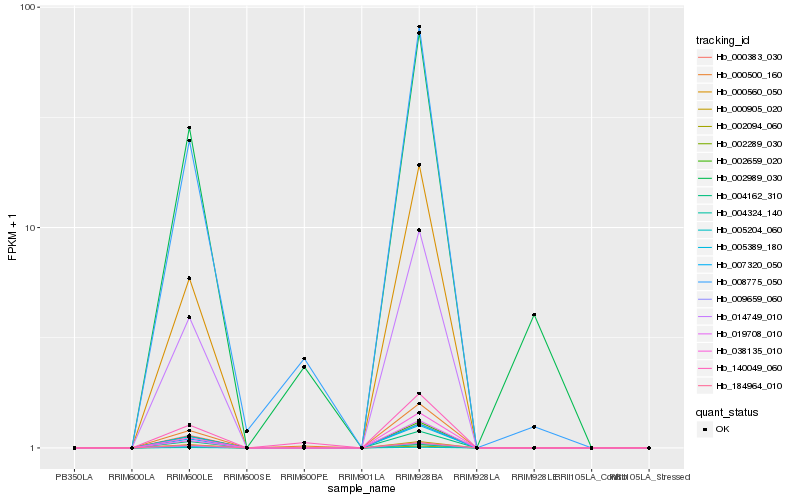
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002989_030 |
0.0 |
- |
- |
PREDICTED: protein GAST1-like [Populus euphratica] |
| 2 |
Hb_008775_050 |
0.1083732619 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 3 |
Hb_000500_160 |
0.1250906779 |
- |
- |
hypothetical protein CISIN_1g039640mg, partial [Citrus sinensis] |
| 4 |
Hb_004162_310 |
0.1435680748 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 5 |
Hb_007320_050 |
0.1437535212 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 6 |
Hb_014749_010 |
0.1443336593 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 7 |
Hb_002659_020 |
0.1455070283 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 8 |
Hb_009659_060 |
0.1458479217 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 9 |
Hb_005389_180 |
0.1530991466 |
- |
- |
PREDICTED: uncharacterized protein LOC105762407 [Gossypium raimondii] |
| 10 |
Hb_000560_050 |
0.1535295479 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 11 |
Hb_000383_030 |
0.1537759646 |
- |
- |
- |
| 12 |
Hb_140049_060 |
0.1540838089 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 13 |
Hb_000905_020 |
0.1574053389 |
- |
- |
PREDICTED: cytochrome P450 93A2-like [Jatropha curcas] |
| 14 |
Hb_038135_010 |
0.158353884 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 15 |
Hb_004324_140 |
0.1587138333 |
- |
- |
PREDICTED: uncharacterized protein LOC105648358 [Jatropha curcas] |
| 16 |
Hb_184964_010 |
0.1590193079 |
- |
- |
- |
| 17 |
Hb_002094_060 |
0.1594318045 |
- |
- |
- |
| 18 |
Hb_005204_060 |
0.1599573811 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_019708_010 |
0.1600820779 |
- |
- |
- |
| 20 |
Hb_002289_030 |
0.1637507732 |
- |
- |
PREDICTED: uncharacterized protein LOC103957017 [Pyrus x bretschneideri] |