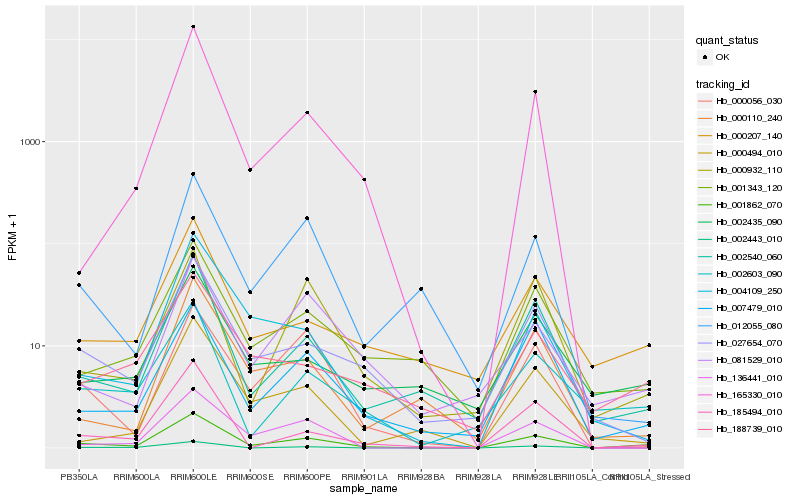
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_070 |
0.0 |
- |
- |
hypothetical protein RCOM_1155060 [Ricinus communis] |
| 2 |
Hb_002540_060 |
0.1736748664 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002603_090 |
0.1863971708 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 4 |
Hb_027654_070 |
0.1954022442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000207_140 |
0.1980824999 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 6 |
Hb_001343_120 |
0.2009514301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_185494_010 |
0.2021932603 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_007479_010 |
0.2036608774 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 9 |
Hb_136441_010 |
0.2041536063 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 10 |
Hb_165330_010 |
0.2073340481 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 11 |
Hb_002443_010 |
0.2109517738 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 12 |
Hb_000932_110 |
0.2119352289 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 13 |
Hb_000056_030 |
0.21401853 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 14 |
Hb_188739_010 |
0.2143804154 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_081529_010 |
0.2156851426 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002435_090 |
0.2172736602 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004109_250 |
0.2184827381 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 18 |
Hb_012055_080 |
0.219334373 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 19 |
Hb_000110_240 |
0.2200740969 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000494_010 |
0.2217087119 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |