| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
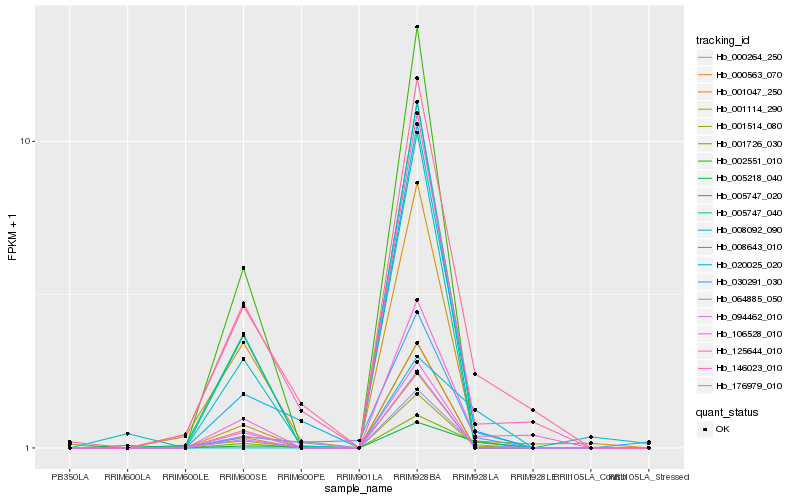
Hb_001726_030 |
0.0 |
- |
- |
Ferulic acid 5-hydroxylase 1 [Theobroma cacao] |
| 2 |
Hb_125644_010 |
0.1961500121 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 3 |
Hb_176979_010 |
0.1992004257 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 4 |
Hb_064885_050 |
0.2304927125 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Populus euphratica] |
| 5 |
Hb_008092_090 |
0.2365387931 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 6 |
Hb_005218_040 |
0.2404872994 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 7 |
Hb_020025_020 |
0.2437851516 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 8 |
Hb_030291_030 |
0.2454359834 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_146023_010 |
0.2481188838 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001047_250 |
0.2526771994 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002551_010 |
0.2535483188 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000264_250 |
0.2536581247 |
- |
- |
thioredoxin family protein [Arabidopsis thaliana] |
| 13 |
Hb_106528_010 |
0.2538170456 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_005747_040 |
0.2538901009 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |
| 15 |
Hb_001514_080 |
0.2539577686 |
- |
- |
hypothetical protein JCGZ_16082 [Jatropha curcas] |
| 16 |
Hb_000563_070 |
0.2541818894 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Sesamum indicum] |
| 17 |
Hb_008643_010 |
0.2545130926 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 18 |
Hb_005747_020 |
0.2553360118 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |
| 19 |
Hb_001114_290 |
0.2555098215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_094462_010 |
0.257412041 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |