| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
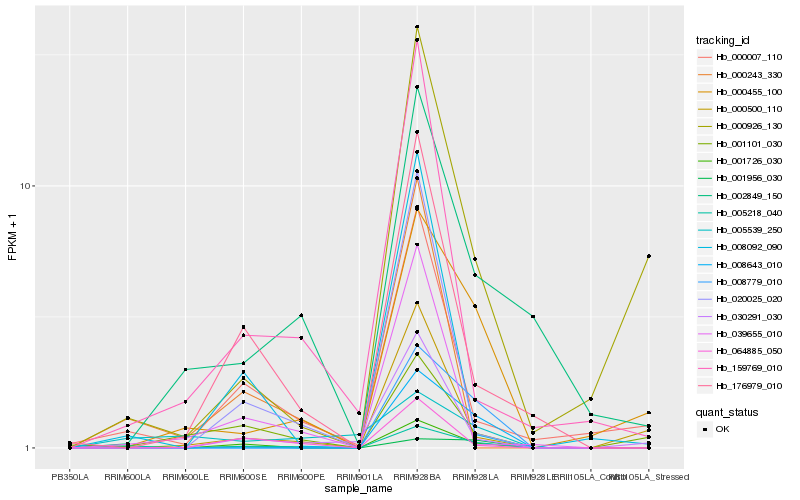
Hb_005218_040 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 2 |
Hb_001726_030 |
0.2404872994 |
- |
- |
Ferulic acid 5-hydroxylase 1 [Theobroma cacao] |
| 3 |
Hb_000455_100 |
0.2658497998 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_008643_010 |
0.2825501271 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 5 |
Hb_000007_110 |
0.2833951943 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 6 |
Hb_008779_010 |
0.2851975396 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 7 |
Hb_176979_010 |
0.2891632901 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 8 |
Hb_064885_050 |
0.2908045603 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Populus euphratica] |
| 9 |
Hb_002849_150 |
0.3014032681 |
- |
- |
hypothetical protein JCGZ_14367 [Jatropha curcas] |
| 10 |
Hb_030291_030 |
0.3014097089 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 11 |
Hb_000926_130 |
0.3119008455 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_159769_010 |
0.3134901331 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_000243_330 |
0.3149141902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_020025_020 |
0.3149856719 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 15 |
Hb_001101_030 |
0.3182153964 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 16 |
Hb_000500_110 |
0.3241581191 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 17 |
Hb_005539_250 |
0.3251750539 |
- |
- |
- |
| 18 |
Hb_001956_030 |
0.3257772766 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 19 |
Hb_039655_010 |
0.3258886267 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 20 |
Hb_008092_090 |
0.3266915444 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |