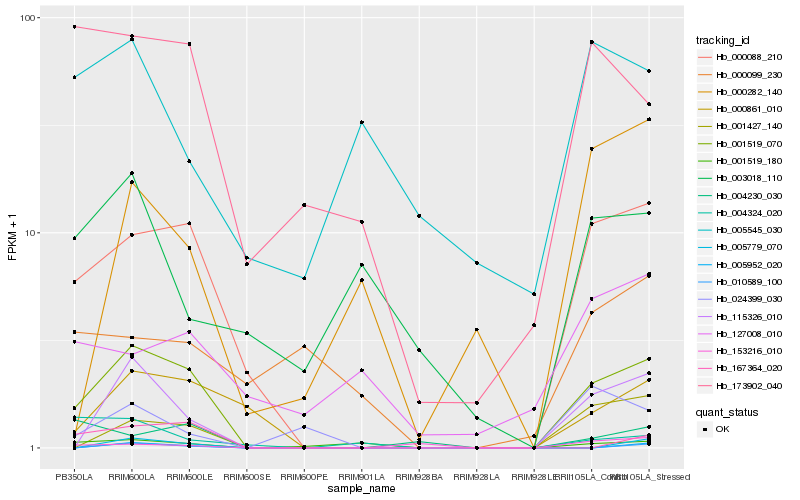
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001519_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_09841 [Jatropha curcas] |
| 2 |
Hb_000088_210 |
0.1695671028 |
- |
- |
hypothetical protein POPTR_0013s11310g [Populus trichocarpa] |
| 3 |
Hb_115326_010 |
0.258421144 |
- |
- |
- |
| 4 |
Hb_000861_010 |
0.268335366 |
- |
- |
- |
| 5 |
Hb_001427_140 |
0.2921042635 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |
| 6 |
Hb_173902_040 |
0.3179499757 |
- |
- |
- |
| 7 |
Hb_167364_020 |
0.3333504448 |
- |
- |
- |
| 8 |
Hb_001519_180 |
0.3375932061 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 9 |
Hb_004230_030 |
0.3394927523 |
- |
- |
hypothetical protein CISIN_1g043871mg [Citrus sinensis] |
| 10 |
Hb_024399_030 |
0.3429121933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_153216_010 |
0.3657781029 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 12 |
Hb_000282_140 |
0.3686558263 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 13 |
Hb_005779_070 |
0.3721670574 |
- |
- |
- |
| 14 |
Hb_005952_020 |
0.3727501557 |
- |
- |
- |
| 15 |
Hb_010589_100 |
0.3747746216 |
- |
- |
Thioredoxin domain-containing protein 9-like protein [Morus notabilis] |
| 16 |
Hb_004324_020 |
0.3770676028 |
- |
- |
- |
| 17 |
Hb_003018_110 |
0.3774992482 |
- |
- |
- |
| 18 |
Hb_127008_010 |
0.3791183014 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 19 |
Hb_005545_030 |
0.3833702353 |
- |
- |
PREDICTED: uncharacterized protein LOC105645697 [Jatropha curcas] |
| 20 |
Hb_000099_230 |
0.3835863094 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |