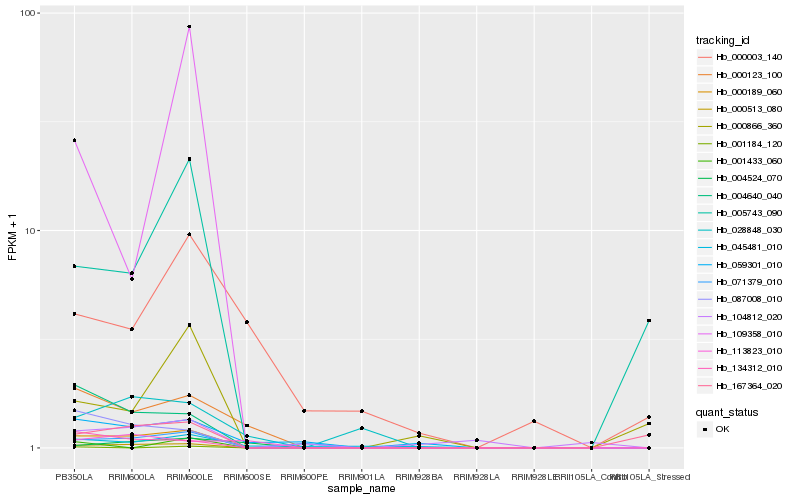
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637477 [Jatropha curcas] |
| 2 |
Hb_000513_080 |
0.0545548477 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 43 [Jatropha curcas] |
| 3 |
Hb_004640_040 |
0.2167238375 |
- |
- |
- |
| 4 |
Hb_134312_010 |
0.2216435379 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 5 |
Hb_087008_010 |
0.2248381897 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 6 |
Hb_071379_010 |
0.2479012236 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 3-like [Nelumbo nucifera] |
| 7 |
Hb_005743_090 |
0.2708600106 |
- |
- |
- |
| 8 |
Hb_000123_100 |
0.2715275796 |
- |
- |
Uncharacterized protein TCM_024608 [Theobroma cacao] |
| 9 |
Hb_059301_010 |
0.2973933392 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 10 |
Hb_167364_020 |
0.309536539 |
- |
- |
- |
| 11 |
Hb_109358_010 |
0.3204046529 |
- |
- |
- |
| 12 |
Hb_045481_010 |
0.3239160679 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000866_360 |
0.3253019305 |
- |
- |
- |
| 14 |
Hb_028848_030 |
0.3302952575 |
- |
- |
- |
| 15 |
Hb_104812_020 |
0.3489152836 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004524_070 |
0.3750190492 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 17 |
Hb_113823_010 |
0.3861691112 |
- |
- |
- |
| 18 |
Hb_000003_140 |
0.3918456447 |
- |
- |
hypothetical protein glysoja_047034, partial [Glycine soja] |
| 19 |
Hb_001184_120 |
0.3964192954 |
- |
- |
hypothetical protein CICLE_v10025281mg [Citrus clementina] |
| 20 |
Hb_001433_060 |
0.3965017049 |
- |
- |
hypothetical protein JCGZ_23291 [Jatropha curcas] |