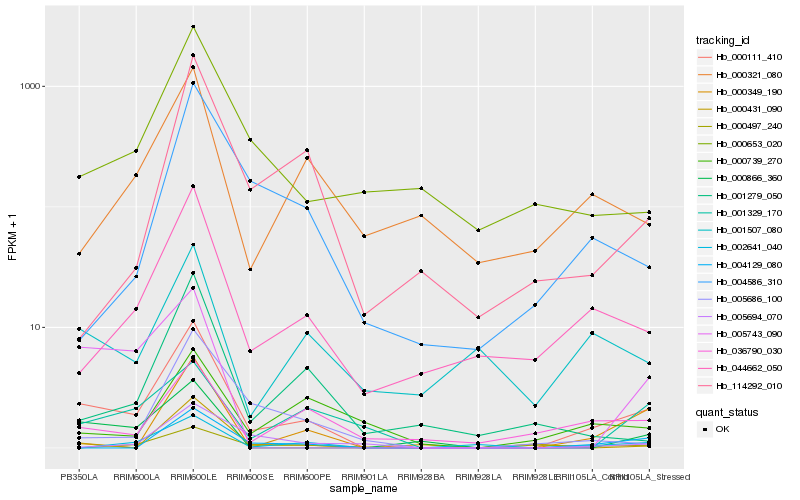
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_410 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_044662_050 |
0.2482298057 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 3 |
Hb_002641_040 |
0.2775436994 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 4 |
Hb_005743_090 |
0.2830812823 |
- |
- |
- |
| 5 |
Hb_000653_020 |
0.2832328577 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 6 |
Hb_000866_360 |
0.2840113429 |
- |
- |
- |
| 7 |
Hb_036790_030 |
0.2866346013 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 8 |
Hb_000349_190 |
0.2901545713 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 9 |
Hb_005686_100 |
0.2918919076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004129_080 |
0.2921978664 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 11 |
Hb_001279_050 |
0.2927111471 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 12 |
Hb_000321_080 |
0.2945292238 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_004586_310 |
0.2949587083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001507_080 |
0.2949791659 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 15 |
Hb_000497_240 |
0.2983909709 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 16 |
Hb_005694_070 |
0.2986713223 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_114292_010 |
0.2991957448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001329_170 |
0.3015036952 |
- |
- |
Chlorophyll a-b binding protein, chloroplastic [Aegilops tauschii] |
| 19 |
Hb_000739_270 |
0.3027605415 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 20 |
Hb_000431_090 |
0.3029274902 |
- |
- |
hypothetical protein POPTR_0017s14020g [Populus trichocarpa] |