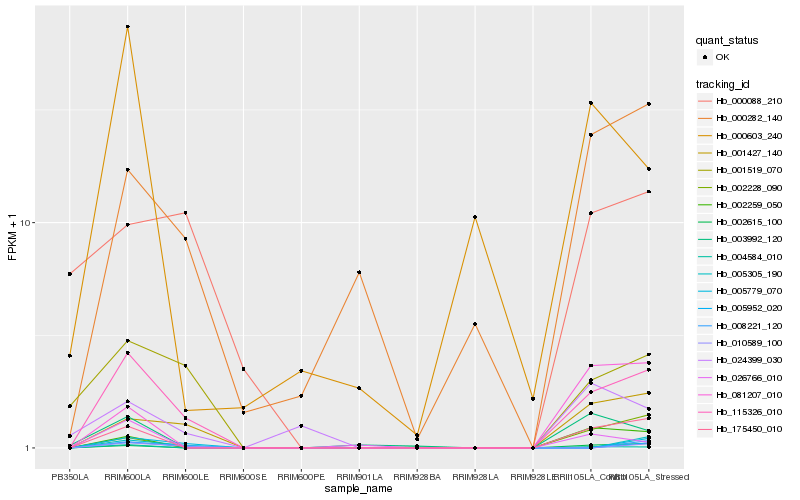
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115326_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001427_140 |
0.2218079385 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |
| 3 |
Hb_175450_010 |
0.2511721818 |
- |
- |
- |
| 4 |
Hb_001519_070 |
0.258421144 |
- |
- |
hypothetical protein JCGZ_09841 [Jatropha curcas] |
| 5 |
Hb_000282_140 |
0.2987232749 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 6 |
Hb_002259_050 |
0.3088753532 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_004584_010 |
0.3179111697 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 8 |
Hb_010589_100 |
0.3280494443 |
- |
- |
Thioredoxin domain-containing protein 9-like protein [Morus notabilis] |
| 9 |
Hb_005952_020 |
0.3317896642 |
- |
- |
- |
| 10 |
Hb_005779_070 |
0.3327951215 |
- |
- |
- |
| 11 |
Hb_008221_120 |
0.3344817933 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g63170-like [Jatropha curcas] |
| 12 |
Hb_002615_100 |
0.3388210561 |
- |
- |
- |
| 13 |
Hb_000088_210 |
0.3405637644 |
- |
- |
hypothetical protein POPTR_0013s11310g [Populus trichocarpa] |
| 14 |
Hb_005305_190 |
0.3448568227 |
transcription factor |
TF Family: NAC |
- |
| 15 |
Hb_003992_120 |
0.3464081804 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 16 |
Hb_081207_010 |
0.3478353487 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 17 |
Hb_002228_090 |
0.3486125549 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 18 |
Hb_026766_010 |
0.349775656 |
- |
- |
- |
| 19 |
Hb_024399_030 |
0.3510184445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000603_240 |
0.3566549131 |
- |
- |
- |