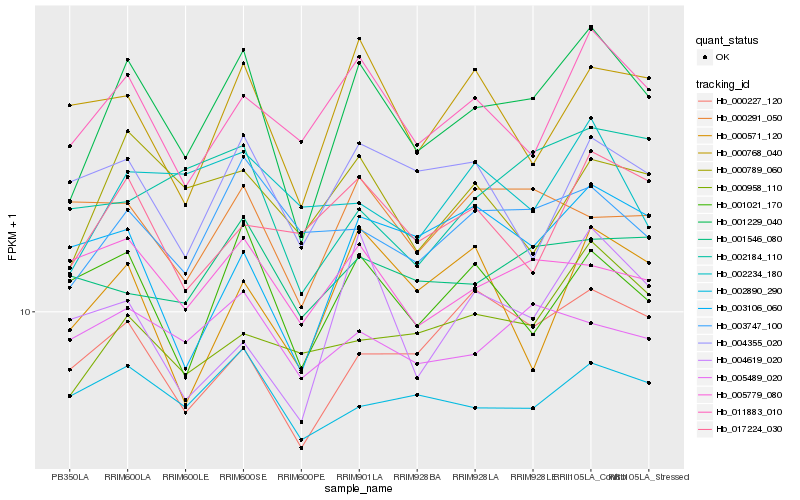
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001229_040 |
0.0 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 2 |
Hb_011883_010 |
0.1073439812 |
- |
- |
PREDICTED: RNA-binding protein 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000789_060 |
0.1171211398 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 6 [Jatropha curcas] |
| 4 |
Hb_001021_170 |
0.1184645343 |
- |
- |
PREDICTED: uncharacterized protein LOC105642474 [Jatropha curcas] |
| 5 |
Hb_003747_100 |
0.1209402063 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000227_120 |
0.1221577706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000958_110 |
0.1248104981 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000768_040 |
0.1255267337 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 9 |
Hb_002890_290 |
0.1264428117 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 10 |
Hb_017224_030 |
0.1277047285 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 11 |
Hb_005779_080 |
0.1297642504 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005489_020 |
0.1309703785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004355_020 |
0.1310918313 |
- |
- |
PREDICTED: lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 14 |
Hb_004619_020 |
0.1312463781 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003106_060 |
0.1315673116 |
- |
- |
hypothetical protein POPTR_0015s08140g [Populus trichocarpa] |
| 16 |
Hb_002234_180 |
0.1319044052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001546_080 |
0.1322311822 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 18 |
Hb_002184_110 |
0.1324825442 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 19 |
Hb_000571_120 |
0.1328951566 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 20 |
Hb_000291_050 |
0.1347830174 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |