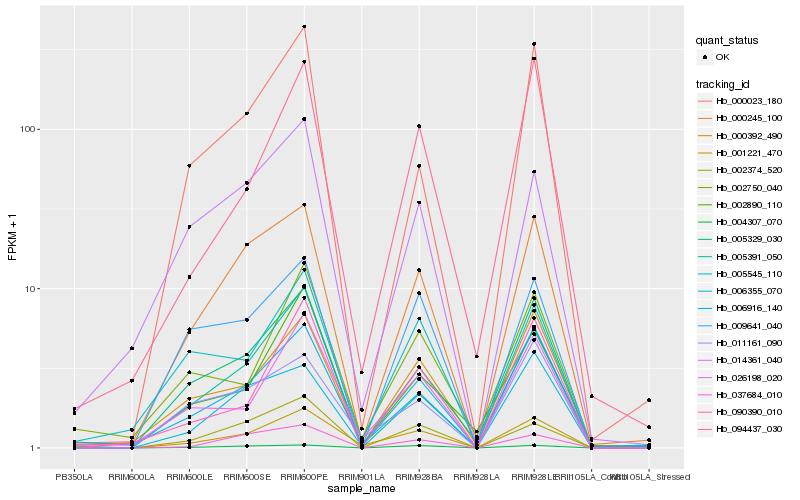
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 2 |
Hb_026198_020 |
0.1240560742 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 3 |
Hb_000245_100 |
0.1312946162 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 4 |
Hb_005329_030 |
0.1338079537 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 5 |
Hb_006916_140 |
0.1401975689 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 6 |
Hb_000392_490 |
0.1457699948 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006355_070 |
0.1499315613 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 8 |
Hb_011161_090 |
0.1549063784 |
- |
- |
- |
| 9 |
Hb_090390_010 |
0.1600318131 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 10 |
Hb_002374_520 |
0.1620831255 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |
| 11 |
Hb_094437_030 |
0.1640146269 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 12 |
Hb_005391_050 |
0.1641100731 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 13 |
Hb_009641_040 |
0.1656051988 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 14 |
Hb_000023_180 |
0.1683594908 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 15 |
Hb_005545_110 |
0.1685543166 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 16 |
Hb_037684_010 |
0.1689357329 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 17 |
Hb_002890_110 |
0.1691251441 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 18 |
Hb_002750_040 |
0.1713043346 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 19 |
Hb_014361_040 |
0.1716957089 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_004307_070 |
0.1717571568 |
- |
- |
kinase, putative [Ricinus communis] |