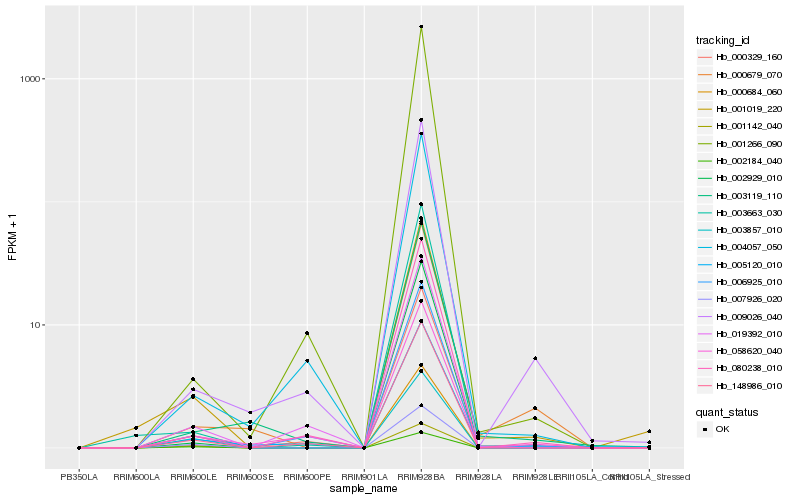
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_220 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s19470g [Populus trichocarpa] |
| 2 |
Hb_000329_160 |
0.1000298191 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000684_060 |
0.1027548682 |
- |
- |
hypothetical protein POPTR_0006s13170g [Populus trichocarpa] |
| 4 |
Hb_003663_030 |
0.1055575627 |
- |
- |
PREDICTED: early nodulin-93-like [Jatropha curcas] |
| 5 |
Hb_005120_010 |
0.1079875059 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_148986_010 |
0.1134330567 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_007926_020 |
0.1140087457 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 8 |
Hb_006925_010 |
0.1160189806 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 9 |
Hb_080238_010 |
0.1161857987 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_002929_010 |
0.1181836439 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 11 |
Hb_003857_010 |
0.1192008322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003119_110 |
0.1205984431 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 13 |
Hb_000679_070 |
0.1224542668 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 14 |
Hb_009026_040 |
0.1227063445 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 15 |
Hb_019392_010 |
0.1233007526 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 16 |
Hb_001142_040 |
0.1234058163 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 17 |
Hb_002184_040 |
0.1243908906 |
transcription factor |
TF Family: bHLH |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004057_050 |
0.1299293179 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 19 |
Hb_001266_090 |
0.1313071122 |
- |
- |
- |
| 20 |
Hb_058620_040 |
0.1317423174 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |