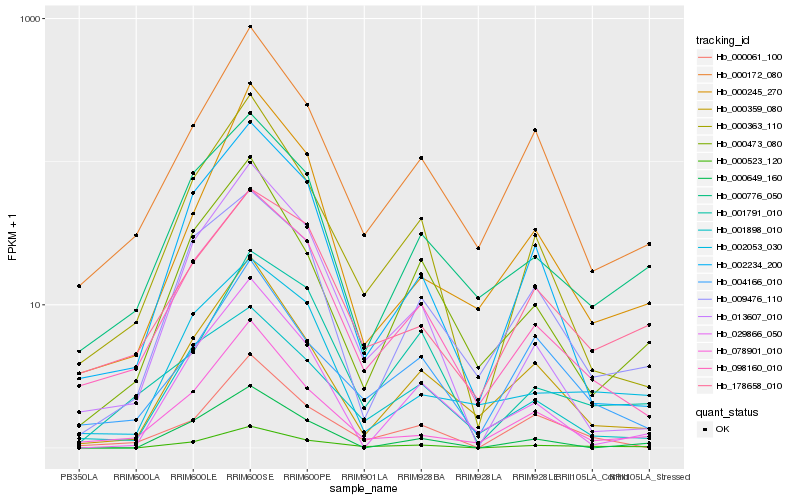
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000523_120 |
0.0 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |
| 2 |
Hb_000363_110 |
0.1632200344 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 21 [Jatropha curcas] |
| 3 |
Hb_000359_080 |
0.1651094136 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 4 |
Hb_098160_010 |
0.1734759914 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000172_080 |
0.177313388 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 6 |
Hb_013607_010 |
0.1774632979 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 7 |
Hb_000473_080 |
0.1780186455 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 8 |
Hb_000649_160 |
0.1782503142 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 9 |
Hb_078901_010 |
0.1786978516 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_002053_030 |
0.1811451638 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002234_200 |
0.181429021 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 12 |
Hb_001898_010 |
0.1819322334 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 13 |
Hb_000061_100 |
0.1821773678 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 14 |
Hb_000776_050 |
0.1826883599 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_009476_110 |
0.1835448713 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 16 |
Hb_000245_270 |
0.185337062 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 17 |
Hb_029866_050 |
0.1873591135 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 18 |
Hb_178658_010 |
0.1889666448 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 19 |
Hb_001791_010 |
0.189051043 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g63710 [Jatropha curcas] |
| 20 |
Hb_004166_010 |
0.1892879988 |
- |
- |
PREDICTED: uncharacterized protein LOC105638508 [Jatropha curcas] |