| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
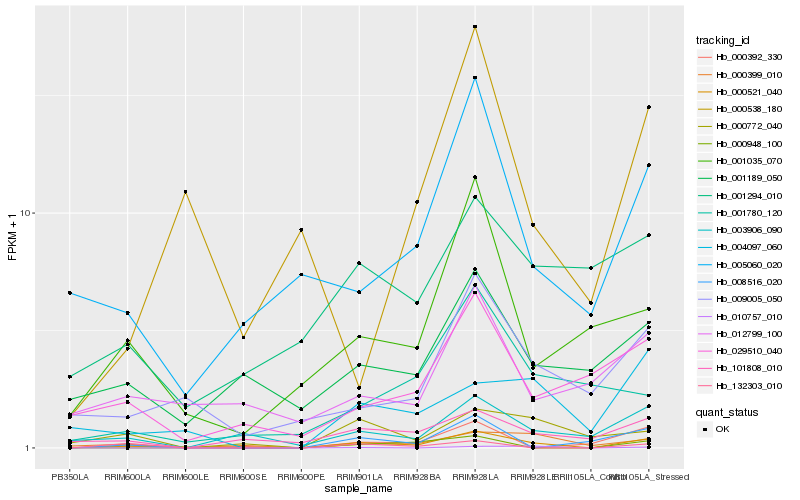
Hb_000521_040 |
0.0 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 2 |
Hb_009005_050 |
0.2947716667 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 3 |
Hb_004097_060 |
0.300392723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001035_070 |
0.3231655597 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 5 |
Hb_005060_020 |
0.3231857479 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 6 |
Hb_003906_090 |
0.3309416174 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 7 |
Hb_000399_010 |
0.3311273255 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 8 |
Hb_008516_020 |
0.3337435046 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 9 |
Hb_101808_010 |
0.3376249102 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000948_100 |
0.3386072418 |
- |
- |
- |
| 11 |
Hb_029510_040 |
0.3394082722 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 12 |
Hb_001780_120 |
0.3398108888 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 13 |
Hb_000392_330 |
0.3440558632 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000772_040 |
0.3444819816 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 15 |
Hb_012799_100 |
0.3456336598 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 16 |
Hb_001294_010 |
0.3492643476 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 17 |
Hb_000538_180 |
0.34936841 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 18 |
Hb_001189_050 |
0.3504680989 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 19 |
Hb_132303_010 |
0.3513891999 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_010757_010 |
0.3523721956 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |