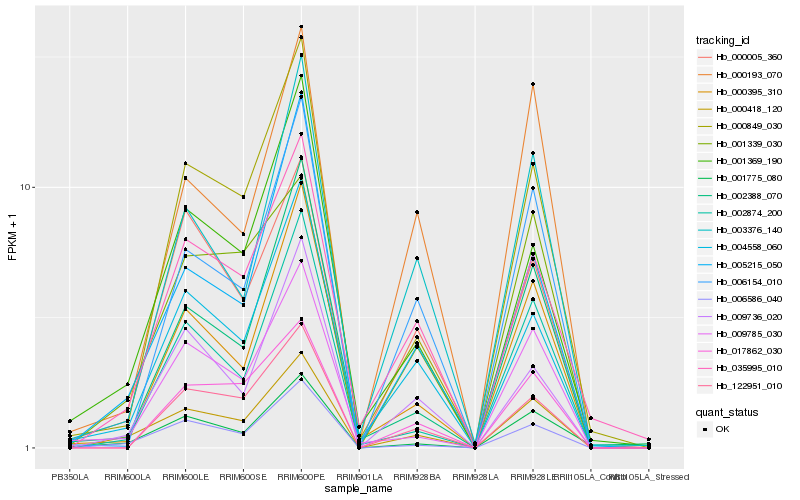
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_120 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 2 |
Hb_006586_040 |
0.1022511813 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |
| 3 |
Hb_001339_030 |
0.1330967045 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 4 |
Hb_004558_060 |
0.1374778076 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 5 |
Hb_035995_010 |
0.1382824261 |
- |
- |
- |
| 6 |
Hb_000849_030 |
0.1421025001 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 7 |
Hb_009785_030 |
0.1438326356 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 8 |
Hb_005215_050 |
0.1441988061 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 9 |
Hb_001369_190 |
0.1472232678 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 10 |
Hb_002874_200 |
0.1472369859 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 11 |
Hb_009736_020 |
0.1489583282 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 12 |
Hb_000395_310 |
0.1491301157 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_006154_010 |
0.1532332879 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000005_360 |
0.1534628969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002388_070 |
0.1555341789 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_003376_140 |
0.1569679807 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 17 |
Hb_001775_080 |
0.1572526913 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 18 |
Hb_000193_070 |
0.1581378841 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 19 |
Hb_017862_030 |
0.1586239429 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 20 |
Hb_122951_010 |
0.158866523 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |