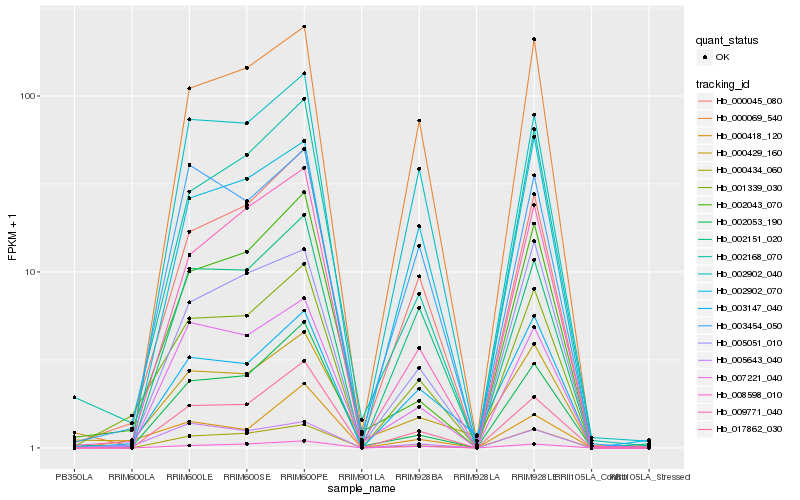
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001339_030 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 2 |
Hb_000045_080 |
0.0959775588 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_002168_070 |
0.1212436463 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_017862_030 |
0.1221689684 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 5 |
Hb_000069_540 |
0.1251788113 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 6 |
Hb_000434_060 |
0.1273823076 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_009771_040 |
0.1283709466 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 8 |
Hb_005051_010 |
0.1292243585 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002043_070 |
0.1297260837 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 10 |
Hb_008598_010 |
0.130044039 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 11 |
Hb_005643_040 |
0.1307968847 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_002151_020 |
0.1322667968 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 13 |
Hb_000418_120 |
0.1330967045 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 14 |
Hb_002053_190 |
0.1334882305 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002902_040 |
0.1343863116 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 16 |
Hb_007221_040 |
0.1373507307 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 17 |
Hb_003454_050 |
0.1381100306 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 18 |
Hb_000429_160 |
0.1385211043 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 19 |
Hb_002902_070 |
0.1388562071 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 20 |
Hb_003147_040 |
0.1389154875 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |