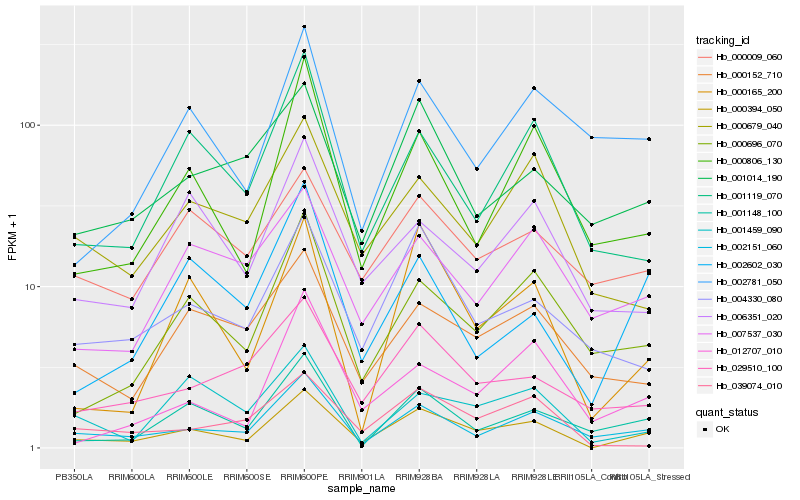
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000394_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_002151_060 |
0.1677623721 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 3 |
Hb_000165_200 |
0.1810675514 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000696_070 |
0.1884108395 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001148_100 |
0.1906720301 |
- |
- |
- |
| 6 |
Hb_000806_130 |
0.1915451717 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 7 |
Hb_004330_080 |
0.191853006 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001119_070 |
0.2016643165 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 9 |
Hb_029510_100 |
0.2051867165 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 10 |
Hb_000152_710 |
0.2057324295 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 11 |
Hb_001014_190 |
0.2086168091 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 12 |
Hb_001459_090 |
0.2086353647 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000679_040 |
0.2095570313 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 14 |
Hb_039074_010 |
0.2096376043 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_012707_010 |
0.2118568618 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 16 |
Hb_007537_030 |
0.2123300175 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000009_060 |
0.212930491 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 18 |
Hb_006351_020 |
0.2131216594 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 19 |
Hb_002781_050 |
0.213587022 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 20 |
Hb_002602_030 |
0.2137956026 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |