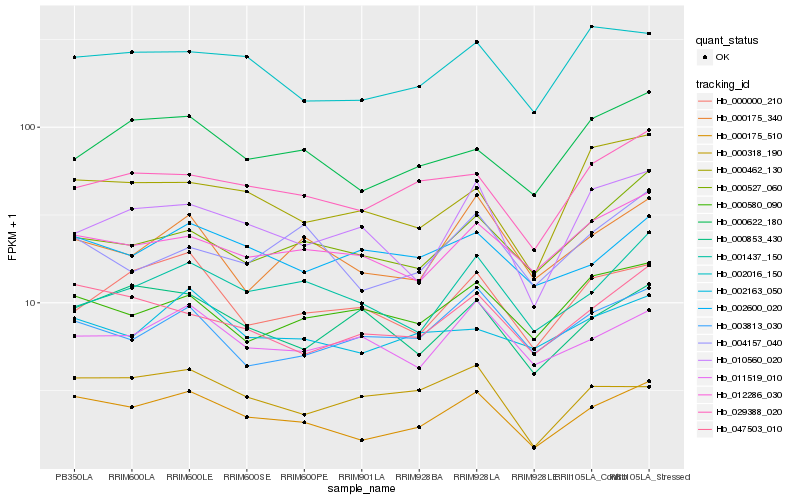
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_510 |
0.0 |
- |
- |
PREDICTED: probable DNA replication complex GINS protein PSF1 [Jatropha curcas] |
| 2 |
Hb_047503_010 |
0.097042519 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 3 |
Hb_002016_150 |
0.1025250147 |
- |
- |
PREDICTED: uncharacterized protein LOC105642512 [Jatropha curcas] |
| 4 |
Hb_029388_020 |
0.1047749614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012286_030 |
0.109333724 |
transcription factor |
TF Family: MED7 |
PREDICTED: mediator of RNA polymerase II transcription subunit 7a [Solanum lycopersicum] |
| 6 |
Hb_000175_340 |
0.1104711603 |
- |
- |
PREDICTED: uncharacterized protein LOC105634861 isoform X3 [Jatropha curcas] |
| 7 |
Hb_002163_050 |
0.112561367 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |
| 8 |
Hb_000622_180 |
0.1128603312 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 9 |
Hb_011519_010 |
0.1134099216 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 10 |
Hb_002600_020 |
0.115591947 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001437_150 |
0.1166573663 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 12 |
Hb_000000_210 |
0.1175941053 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000527_060 |
0.1176885048 |
- |
- |
Protein C20orf11, putative [Ricinus communis] |
| 14 |
Hb_003813_030 |
0.1183498352 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 15 |
Hb_000462_130 |
0.1186467437 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 16 |
Hb_000318_190 |
0.1202748136 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 17 |
Hb_010560_020 |
0.1208780871 |
- |
- |
PREDICTED: uncharacterized protein LOC105649267 [Jatropha curcas] |
| 18 |
Hb_000580_090 |
0.1219269685 |
- |
- |
PREDICTED: tRNA 2'-phosphotransferase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004157_040 |
0.1221749386 |
- |
- |
Rab2 [Hevea brasiliensis] |
| 20 |
Hb_000853_430 |
0.1225739789 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |