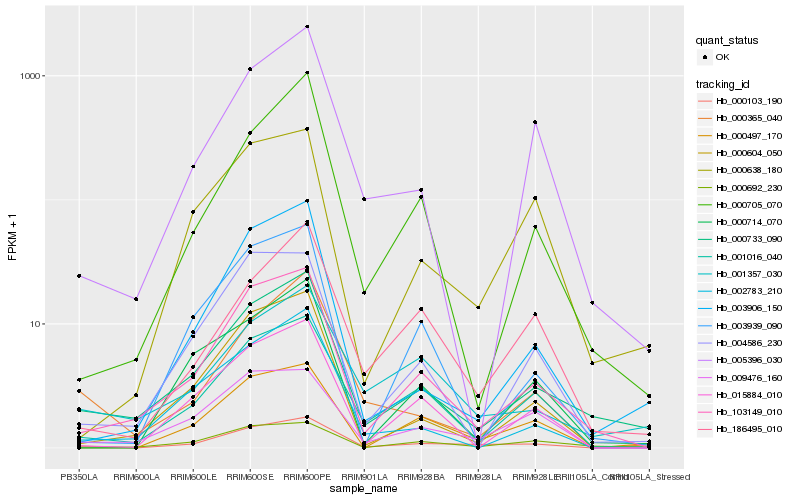
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_190 |
0.0 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 2 |
Hb_001016_040 |
0.1584065981 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_103149_010 |
0.1588772864 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 4 |
Hb_009476_160 |
0.1739010265 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 5 |
Hb_000733_090 |
0.175041146 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 6 |
Hb_000497_170 |
0.1838065944 |
- |
- |
PREDICTED: uncharacterized protein LOC105647275 [Jatropha curcas] |
| 7 |
Hb_015884_010 |
0.1891057599 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000365_040 |
0.1924011456 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 9 |
Hb_000604_050 |
0.1944545473 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 10 |
Hb_001357_030 |
0.1995828231 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 11 |
Hb_000692_230 |
0.2007259191 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 12 |
Hb_186495_010 |
0.2027481993 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 13 |
Hb_004586_230 |
0.205266322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003939_090 |
0.2060187586 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_005396_030 |
0.2061881515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000638_180 |
0.2077784448 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 17 |
Hb_000714_070 |
0.2096304026 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 18 |
Hb_003906_150 |
0.2115482029 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 19 |
Hb_002783_210 |
0.2125715527 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 20 |
Hb_000705_070 |
0.2129196571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |