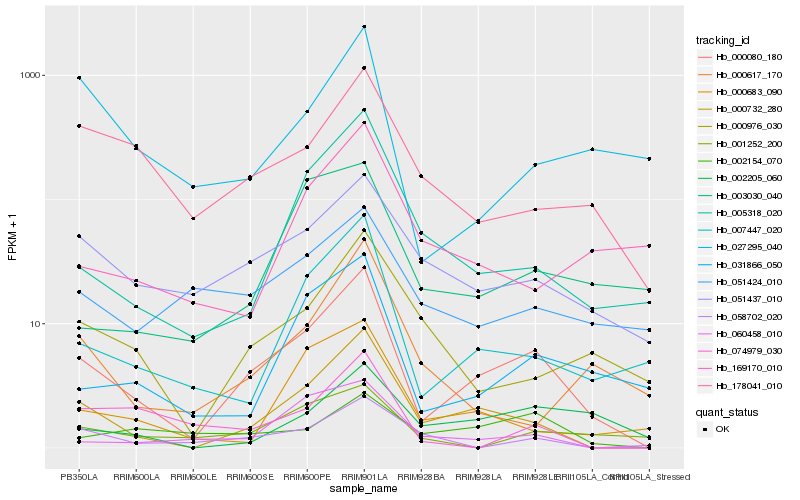
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_000732_280 |
0.1716783894 |
- |
- |
hypothetical protein B456_009G173700 [Gossypium raimondii] |
| 3 |
Hb_007447_020 |
0.2235817566 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 4 |
Hb_031866_050 |
0.2375831237 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 5 |
Hb_051437_010 |
0.2415428937 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005318_020 |
0.2482140101 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_060458_010 |
0.2571195435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000976_030 |
0.263074696 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like [Citrus sinensis] |
| 9 |
Hb_178041_010 |
0.2656929106 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 10 |
Hb_000683_090 |
0.2667647088 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 11 |
Hb_002205_060 |
0.2745417164 |
- |
- |
- |
| 12 |
Hb_051424_010 |
0.280422132 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 13 |
Hb_027295_040 |
0.280907885 |
- |
- |
hypothetical protein MIMGU_mgv1a017500mg [Erythranthe guttata] |
| 14 |
Hb_000617_170 |
0.2827656447 |
- |
- |
- |
| 15 |
Hb_003030_040 |
0.2841411795 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 16 |
Hb_001252_200 |
0.2857627674 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 17 |
Hb_074979_030 |
0.2859462759 |
- |
- |
HSP20-like chaperones superfamily protein [Theobroma cacao] |
| 18 |
Hb_058702_020 |
0.2865879477 |
- |
- |
PREDICTED: uncharacterized protein LOC104604814 [Nelumbo nucifera] |
| 19 |
Hb_002154_070 |
0.2868356705 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 20 |
Hb_169170_010 |
0.2919284837 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |