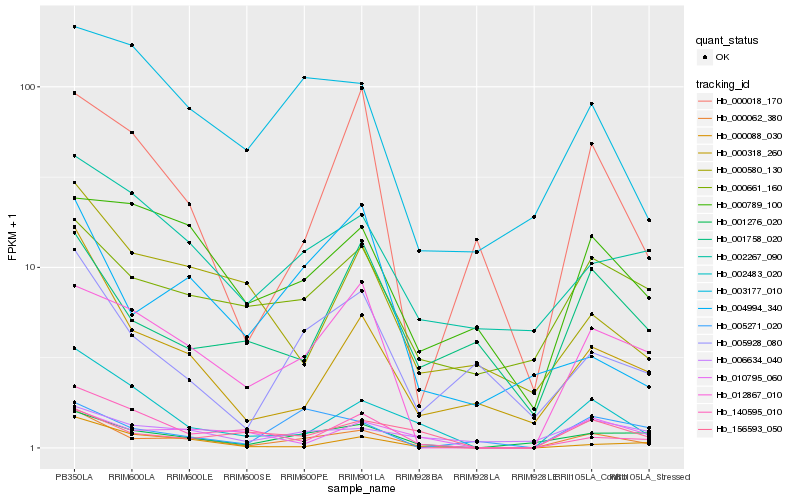
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_380 |
0.0 |
- |
- |
PREDICTED: putative F-box protein At1g67623 [Jatropha curcas] |
| 2 |
Hb_001276_020 |
0.2041872058 |
- |
- |
hypothetical protein VITISV_003263 [Vitis vinifera] |
| 3 |
Hb_002483_020 |
0.2050086271 |
- |
- |
- |
| 4 |
Hb_000318_260 |
0.2079266681 |
- |
- |
PREDICTED: cell division cycle-associated 7-like protein [Jatropha curcas] |
| 5 |
Hb_012867_010 |
0.2189260025 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 6 |
Hb_005928_080 |
0.23187188 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000088_030 |
0.2356907179 |
- |
- |
hypothetical protein JCGZ_10947 [Jatropha curcas] |
| 8 |
Hb_156593_050 |
0.2501819651 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000018_170 |
0.2513572679 |
- |
- |
PREDICTED: bark storage protein A-like [Jatropha curcas] |
| 10 |
Hb_000661_160 |
0.2516879604 |
- |
- |
hexokinase [Manihot esculenta] |
| 11 |
Hb_003177_010 |
0.2525139584 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 12 |
Hb_002267_090 |
0.253188228 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 13 |
Hb_140595_010 |
0.2546150899 |
- |
- |
- |
| 14 |
Hb_010795_060 |
0.2560535623 |
- |
- |
- |
| 15 |
Hb_004994_340 |
0.2578470733 |
- |
- |
- |
| 16 |
Hb_006634_040 |
0.2597310404 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 17 |
Hb_005271_020 |
0.2636542616 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 18 |
Hb_000580_130 |
0.2643555102 |
- |
- |
hypothetical protein JCGZ_14541 [Jatropha curcas] |
| 19 |
Hb_001758_020 |
0.2645342947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000789_100 |
0.2656249676 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |