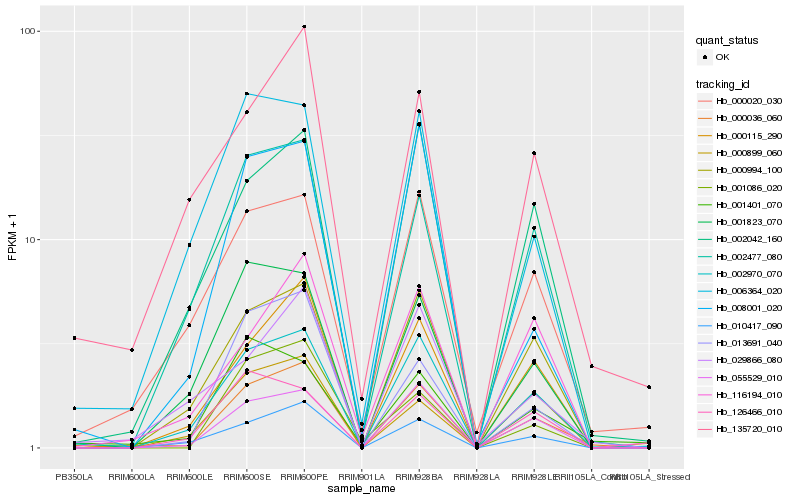
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000036_060 |
0.0 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 2 |
Hb_002970_070 |
0.128410383 |
- |
- |
multidrug/pheromone exporter protein [Hevea brasiliensis] |
| 3 |
Hb_055529_010 |
0.1425965429 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 4 |
Hb_010417_090 |
0.1437419516 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 5 |
Hb_000115_290 |
0.152141178 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 6 |
Hb_000994_100 |
0.1542489796 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 7 |
Hb_013691_040 |
0.1608054103 |
- |
- |
hypothetical protein B456_001G045900 [Gossypium raimondii] |
| 8 |
Hb_002477_080 |
0.1609321493 |
- |
- |
hypothetical protein RCOM_0653450 [Ricinus communis] |
| 9 |
Hb_002042_160 |
0.1659414938 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_000020_030 |
0.1787150825 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001823_070 |
0.1787900466 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 12 |
Hb_001086_020 |
0.1820172465 |
- |
- |
hypothetical protein POPTR_0013s01730g [Populus trichocarpa] |
| 13 |
Hb_000899_060 |
0.1841606498 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 14 |
Hb_116194_010 |
0.1888632323 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_135720_010 |
0.1895029474 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 16 |
Hb_126466_010 |
0.1921673502 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_008001_020 |
0.192531453 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 18 |
Hb_006364_020 |
0.1931937897 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 19 |
Hb_001401_070 |
0.1932273753 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 20 |
Hb_029866_080 |
0.1948174178 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |