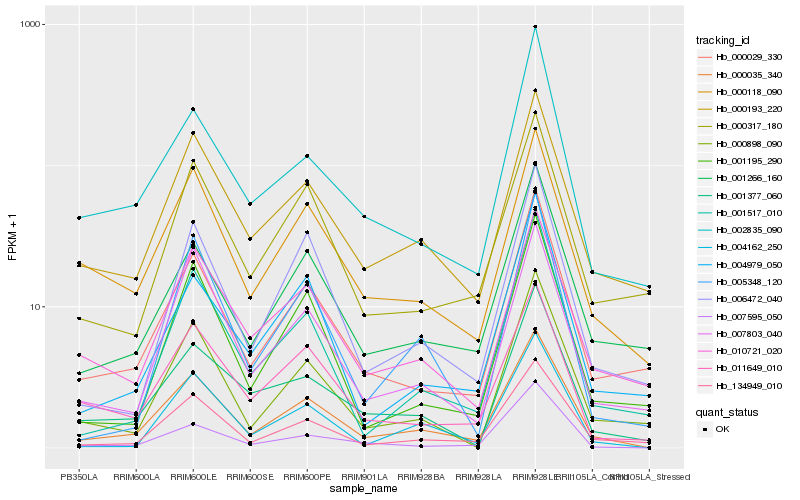
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_134949_010 |
0.1094696371 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 3 |
Hb_000029_330 |
0.122545251 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000193_220 |
0.1232049219 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006472_040 |
0.1274836118 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004162_250 |
0.1277980661 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 7 |
Hb_011649_010 |
0.1295981258 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 8 |
Hb_000118_090 |
0.1297499318 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_007803_040 |
0.1303278965 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001517_010 |
0.1305880304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001266_160 |
0.1322447345 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001377_060 |
0.1343814464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001195_290 |
0.1352144269 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 14 |
Hb_010721_020 |
0.1373682691 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002835_090 |
0.1375217237 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 16 |
Hb_000898_090 |
0.1377225751 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105633600 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000317_180 |
0.138878589 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007595_050 |
0.139276301 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 19 |
Hb_004979_050 |
0.1421506272 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 20 |
Hb_005348_120 |
0.1423386163 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |