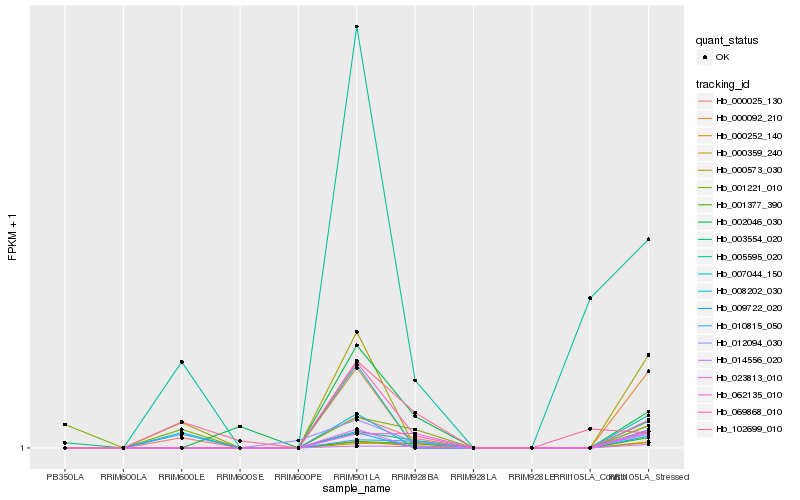
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_010815_050 |
0.2578609912 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 3 |
Hb_000573_030 |
0.2690970379 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 4 |
Hb_000252_140 |
0.2718002942 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_007044_150 |
0.2911097549 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 8A-like [Jatropha curcas] |
| 6 |
Hb_001377_390 |
0.3121592542 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |
| 7 |
Hb_069868_010 |
0.3198603381 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 8 |
Hb_005595_020 |
0.327306299 |
- |
- |
- |
| 9 |
Hb_014556_020 |
0.329310937 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_062135_010 |
0.3309069202 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 11 |
Hb_000359_240 |
0.3353247043 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 12 |
Hb_102699_010 |
0.3506749448 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR type disease resistance protein [Populus trichocarpa] |
| 13 |
Hb_001221_010 |
0.356714544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003554_020 |
0.3706990379 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 15 |
Hb_002046_030 |
0.3720388607 |
- |
- |
PREDICTED: uncharacterized protein LOC104788746 [Camelina sativa] |
| 16 |
Hb_012094_030 |
0.3734514792 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 17 |
Hb_009722_020 |
0.3811598675 |
- |
- |
- |
| 18 |
Hb_023813_010 |
0.3819737981 |
- |
- |
PREDICTED: uncharacterized protein LOC105162193 [Sesamum indicum] |
| 19 |
Hb_000092_210 |
0.3853273973 |
- |
- |
- |
| 20 |
Hb_008202_030 |
0.3854390508 |
- |
- |
hypothetical protein B456_002G115600 [Gossypium raimondii] |