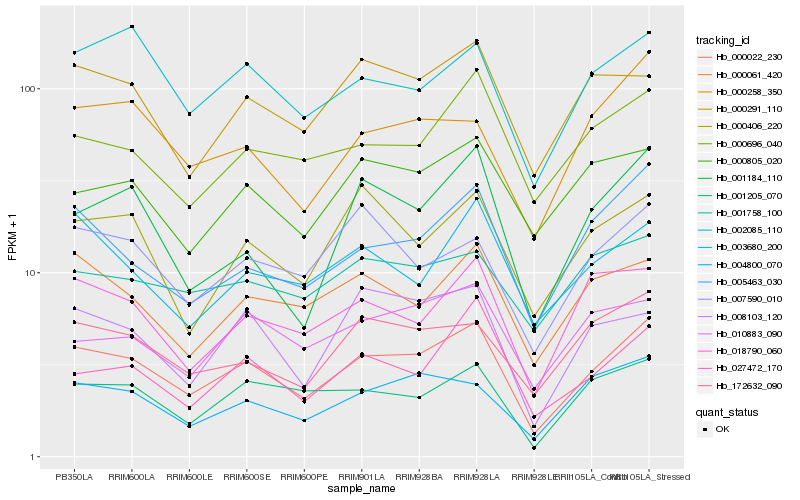
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_230 |
0.0 |
- |
- |
hypothetical protein RCOM_0803470 [Ricinus communis] |
| 2 |
Hb_005463_030 |
0.1110979658 |
- |
- |
PREDICTED: uncharacterized protein LOC105632512 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001205_070 |
0.1126876073 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 4 |
Hb_172632_090 |
0.1149880742 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 5 |
Hb_004800_070 |
0.1151407397 |
- |
- |
- |
| 6 |
Hb_002085_110 |
0.1162016281 |
- |
- |
Protein transport protein Sec61 subunit beta, putative [Ricinus communis] |
| 7 |
Hb_000061_420 |
0.1164577681 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 8 |
Hb_000696_040 |
0.11668147 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_018790_060 |
0.1168646368 |
- |
- |
hypothetical protein JCGZ_21628 [Jatropha curcas] |
| 10 |
Hb_027472_170 |
0.1187674441 |
- |
- |
hypothetical protein POPTR_0007s08080g [Populus trichocarpa] |
| 11 |
Hb_010883_090 |
0.1234187147 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000291_110 |
0.1236154161 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 13 |
Hb_001184_110 |
0.1240529663 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000805_020 |
0.1248245078 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_003680_200 |
0.1249102908 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 16 |
Hb_000258_350 |
0.1252926931 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 17 |
Hb_000406_220 |
0.1264184925 |
- |
- |
hypothetical protein POPTR_0005s04700g [Populus trichocarpa] |
| 18 |
Hb_008103_120 |
0.1267526062 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 19 |
Hb_007590_010 |
0.1277759391 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_001758_100 |
0.1283739424 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |