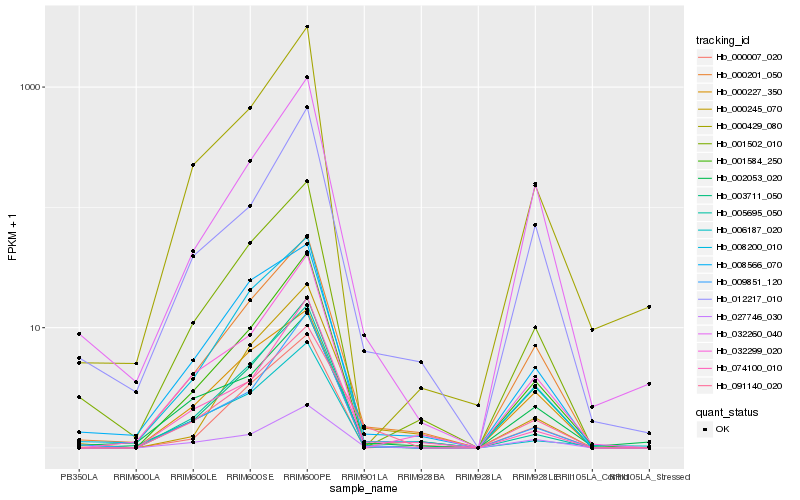
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091140_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640903 [Jatropha curcas] |
| 2 |
Hb_027746_030 |
0.0984048848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000201_050 |
0.1202699815 |
- |
- |
- |
| 4 |
Hb_001584_250 |
0.1393051307 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 5 |
Hb_005695_050 |
0.1474081917 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 6 |
Hb_008200_010 |
0.1488280503 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000227_350 |
0.1534616092 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 8 |
Hb_003711_050 |
0.1542860603 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 9 |
Hb_002053_020 |
0.1553408827 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 10 |
Hb_032260_040 |
0.1563632346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009851_120 |
0.1574495882 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 12 |
Hb_000245_070 |
0.1575107429 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 13 |
Hb_001502_010 |
0.1586161427 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 14 |
Hb_000429_080 |
0.1606277584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012217_010 |
0.1614044836 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 16 |
Hb_074100_010 |
0.1620130305 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 17 |
Hb_008566_070 |
0.1624149923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_032299_020 |
0.162922056 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 19 |
Hb_000007_020 |
0.1652989147 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 20 |
Hb_006187_020 |
0.1667071103 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |