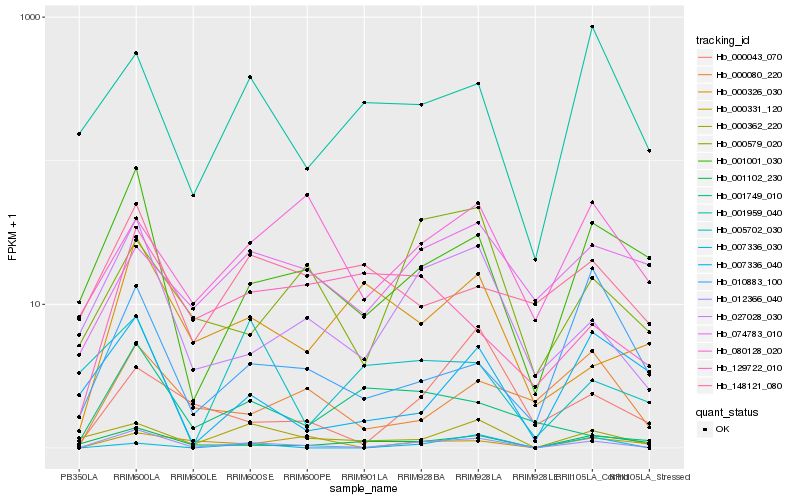
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012366_040 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Gossypium raimondii] |
| 2 |
Hb_027028_030 |
0.2410167592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001001_030 |
0.2560438609 |
- |
- |
hypothetical protein POPTR_0014s03230g [Populus trichocarpa] |
| 4 |
Hb_000362_220 |
0.2878298771 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 5 |
Hb_001102_230 |
0.2953875094 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 1-like [Jatropha curcas] |
| 6 |
Hb_007336_040 |
0.3231884425 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000579_020 |
0.3253530647 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 8 |
Hb_000080_220 |
0.3282763308 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001959_040 |
0.3327776493 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000326_030 |
0.3342289492 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |
| 11 |
Hb_000043_070 |
0.3349196483 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 12 |
Hb_010883_100 |
0.336783288 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 13 |
Hb_000331_120 |
0.3372090629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001749_010 |
0.3511235208 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 15 |
Hb_148121_080 |
0.3530008013 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 16 |
Hb_080128_020 |
0.3566283265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_074783_010 |
0.3595642684 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 18 |
Hb_005702_030 |
0.3619688531 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 19 |
Hb_129722_010 |
0.3632568152 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 20 |
Hb_007336_030 |
0.366368416 |
- |
- |
- |