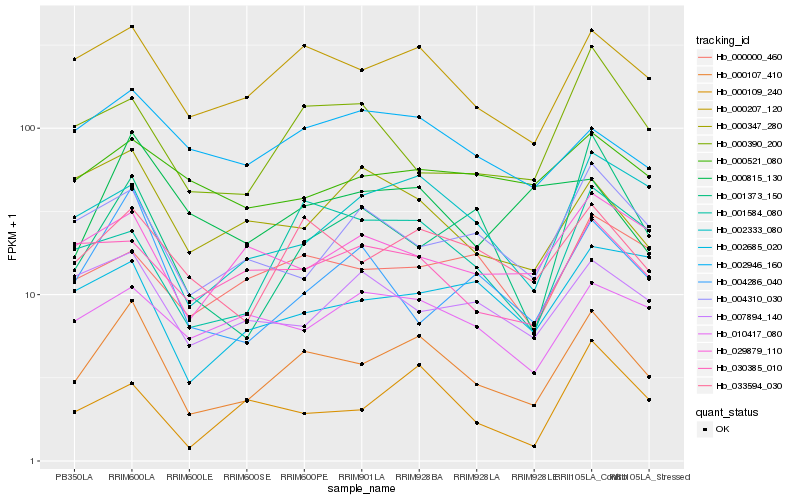
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_410 |
0.0 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 2 |
Hb_000207_120 |
0.1315109912 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 3 |
Hb_033594_030 |
0.136900944 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 4 |
Hb_002333_080 |
0.1688604615 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20-like [Populus euphratica] |
| 5 |
Hb_001584_080 |
0.1796436252 |
- |
- |
hypothetical protein JCGZ_23206 [Jatropha curcas] |
| 6 |
Hb_004286_040 |
0.1827607261 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002946_160 |
0.187932191 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 8 |
Hb_030385_010 |
0.1882212554 |
- |
- |
PREDICTED: zinc-binding alcohol dehydrogenase domain-containing protein 2-like [Jatropha curcas] |
| 9 |
Hb_029879_110 |
0.1882947176 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004310_030 |
0.1894397432 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 11 |
Hb_000109_240 |
0.1908862116 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 12 |
Hb_010417_080 |
0.1915039817 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007894_140 |
0.1915906737 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 14 |
Hb_000815_130 |
0.1920867244 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 15 |
Hb_000000_460 |
0.1921445538 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 16 |
Hb_000347_280 |
0.1926531235 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 17 |
Hb_000390_200 |
0.1943882787 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 18 |
Hb_000521_080 |
0.1961176934 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |
| 19 |
Hb_002685_020 |
0.1963898811 |
- |
- |
- |
| 20 |
Hb_001373_150 |
0.196493807 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |