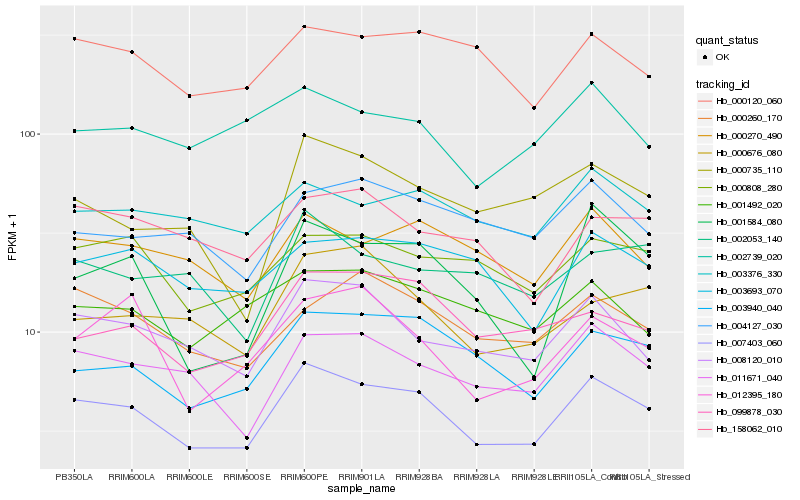
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_060 |
0.0 |
- |
- |
PREDICTED: CAAX prenyl protease 2 [Jatropha curcas] |
| 2 |
Hb_008120_010 |
0.0942014663 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 3 |
Hb_011671_040 |
0.0950040043 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003940_040 |
0.0972964083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000270_490 |
0.1049955491 |
- |
- |
betaine aldehyde dehydrogenase 1 [Jatropha curcas] |
| 6 |
Hb_000808_280 |
0.1079561032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000120_060 |
0.1094272157 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 8 |
Hb_099878_030 |
0.1100214762 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 9 |
Hb_003693_070 |
0.1112944066 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 10 |
Hb_003376_330 |
0.1128013883 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 11 |
Hb_000260_170 |
0.1129673965 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 12 |
Hb_001584_080 |
0.1129767949 |
- |
- |
hypothetical protein JCGZ_23206 [Jatropha curcas] |
| 13 |
Hb_000676_080 |
0.1153756174 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 14 |
Hb_001492_020 |
0.1158608775 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 15 |
Hb_002739_020 |
0.1164794245 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 16 |
Hb_012395_180 |
0.1169840099 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_002053_140 |
0.1170308112 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 18 |
Hb_004127_030 |
0.1179730765 |
- |
- |
putative Rab geranylgeranyl transferase type II beta subunit family protein [Populus trichocarpa] |
| 19 |
Hb_158062_010 |
0.1180628282 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 20 |
Hb_000735_110 |
0.1181337198 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |