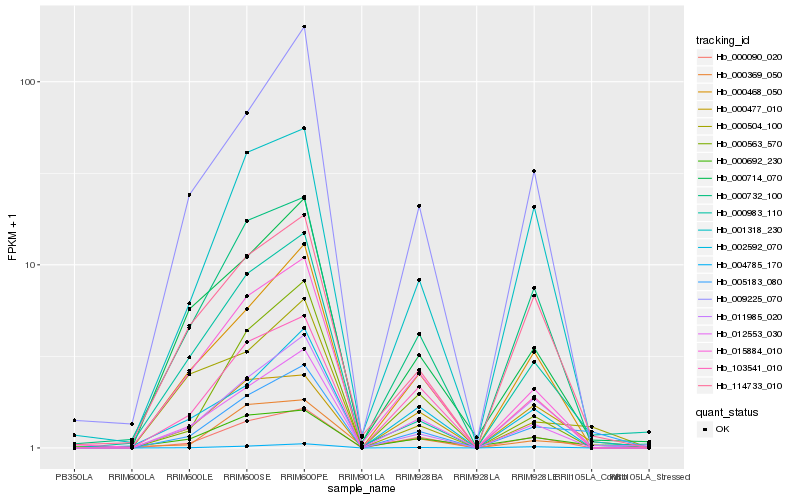
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_011985_020 |
0.1800134065 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_012553_030 |
0.1876591398 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 4 |
Hb_000468_050 |
0.1910273053 |
- |
- |
- |
| 5 |
Hb_000090_020 |
0.1913810691 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000504_100 |
0.1927321134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009225_070 |
0.1984454691 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 8 |
Hb_004785_170 |
0.199796684 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 9 |
Hb_103541_010 |
0.200079488 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000369_050 |
0.2009374771 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 11 |
Hb_000732_100 |
0.2013267124 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_015884_010 |
0.2017028781 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 13 |
Hb_114733_010 |
0.2017244926 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 14 |
Hb_000983_110 |
0.202348228 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 15 |
Hb_000714_070 |
0.2038782501 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 16 |
Hb_000477_010 |
0.2055277514 |
- |
- |
PREDICTED: potassium channel AKT1-like [Jatropha curcas] |
| 17 |
Hb_000563_570 |
0.2065579991 |
- |
- |
hypothetical protein JCGZ_18895 [Jatropha curcas] |
| 18 |
Hb_001318_230 |
0.2068691318 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 19 |
Hb_000692_230 |
0.2072487825 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 20 |
Hb_002592_070 |
0.2075419002 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |