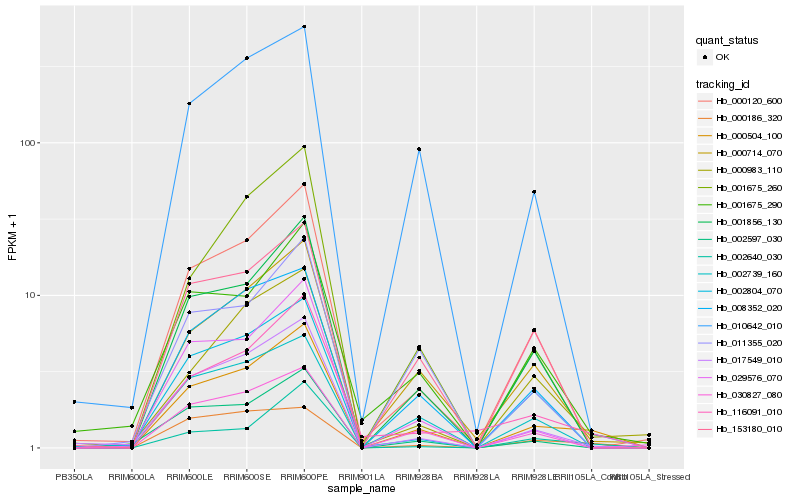
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001856_130 |
0.1341208288 |
- |
- |
hypothetical protein CISIN_1g022795mg [Citrus sinensis] |
| 3 |
Hb_000714_070 |
0.1369659644 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 4 |
Hb_017549_010 |
0.1402644876 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 5 |
Hb_116091_010 |
0.1424886675 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0007s13380g [Populus trichocarpa] |
| 6 |
Hb_000120_600 |
0.1430537882 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 7 |
Hb_030827_080 |
0.1461100206 |
- |
- |
PREDICTED: uncharacterized protein LOC105648491 [Jatropha curcas] |
| 8 |
Hb_008352_020 |
0.1515424495 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 9 |
Hb_029576_070 |
0.1532936212 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 10 |
Hb_153180_010 |
0.155828595 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 11 |
Hb_002739_160 |
0.1576444897 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_010642_010 |
0.1582542523 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 13 |
Hb_002597_030 |
0.1584157677 |
- |
- |
- |
| 14 |
Hb_011355_020 |
0.1623961772 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 15 |
Hb_001675_290 |
0.1639360482 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002640_030 |
0.1654869254 |
- |
- |
hypothetical protein POPTR_0014s05270g [Populus trichocarpa] |
| 17 |
Hb_000186_320 |
0.1669192467 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001675_260 |
0.1686585508 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_000983_110 |
0.1687106518 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 20 |
Hb_002804_070 |
0.1702057251 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |