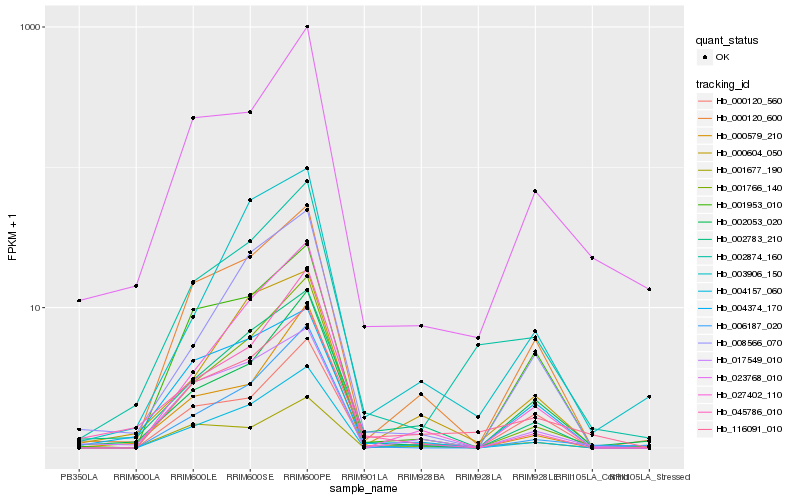
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002874_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_116091_010 |
0.1217443389 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0007s13380g [Populus trichocarpa] |
| 3 |
Hb_023768_010 |
0.1490171418 |
- |
- |
- |
| 4 |
Hb_002053_020 |
0.1607779216 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 5 |
Hb_000120_600 |
0.1633923998 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 6 |
Hb_001766_140 |
0.1706818013 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 7 |
Hb_002783_210 |
0.1751352026 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 8 |
Hb_008566_070 |
0.1762771216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004374_170 |
0.1811674742 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 10 |
Hb_006187_020 |
0.1813429612 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |
| 11 |
Hb_017549_010 |
0.1813640525 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 12 |
Hb_001677_190 |
0.1817074772 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_004157_060 |
0.1835907832 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |
| 14 |
Hb_001953_010 |
0.1845416318 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 15 |
Hb_003906_150 |
0.1849399981 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 16 |
Hb_000604_050 |
0.186402977 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 17 |
Hb_000579_210 |
0.1864142287 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 18 |
Hb_045786_010 |
0.1868710713 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 19 |
Hb_000120_560 |
0.1896807816 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_027402_110 |
0.1906208803 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |