| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
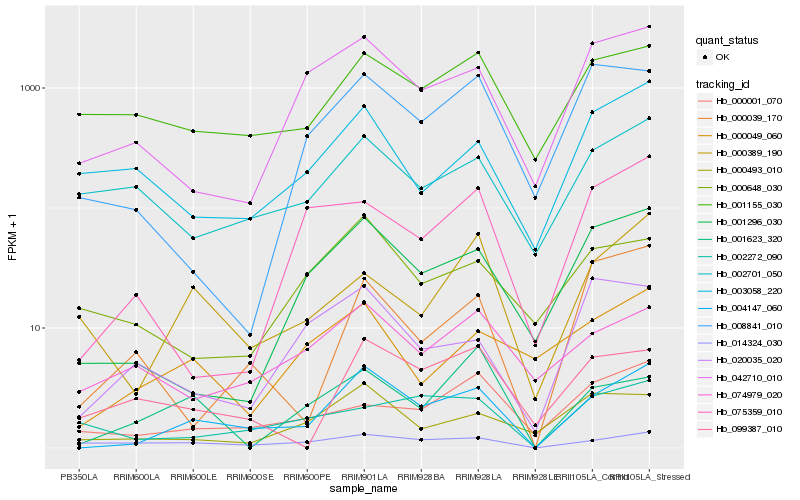
Hb_004147_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 2 |
Hb_001296_030 |
0.2269106165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 3 |
Hb_042710_010 |
0.2388271333 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 4 |
Hb_000493_010 |
0.2506841675 |
- |
- |
hypothetical protein CICLE_v10021431mg [Citrus clementina] |
| 5 |
Hb_014324_030 |
0.2520444187 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 6 |
Hb_000389_190 |
0.2521982693 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A1-like [Jatropha curcas] |
| 7 |
Hb_000049_060 |
0.2524456073 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_074979_020 |
0.257335233 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001155_030 |
0.2585411908 |
- |
- |
60S ribosomal protein L10A [Hevea brasiliensis] |
| 10 |
Hb_000001_070 |
0.2594542913 |
- |
- |
- |
| 11 |
Hb_099387_010 |
0.2598619898 |
- |
- |
- |
| 12 |
Hb_000039_170 |
0.2620985818 |
- |
- |
protein MOTHER of FT and TF 1-like [Jatropha curcas] |
| 13 |
Hb_020035_020 |
0.2678912658 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 14 |
Hb_002701_050 |
0.2693301532 |
- |
- |
PREDICTED: 60S ribosomal protein L7a-like [Jatropha curcas] |
| 15 |
Hb_008841_010 |
0.2694437748 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 16 |
Hb_000648_030 |
0.2703519524 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 17 |
Hb_075359_010 |
0.2708161908 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 18 |
Hb_003058_220 |
0.2713108991 |
- |
- |
Pre-mRNA branch site protein p14, putative [Ricinus communis] |
| 19 |
Hb_002272_090 |
0.2716145351 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 20 |
Hb_001623_320 |
0.2727653916 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |