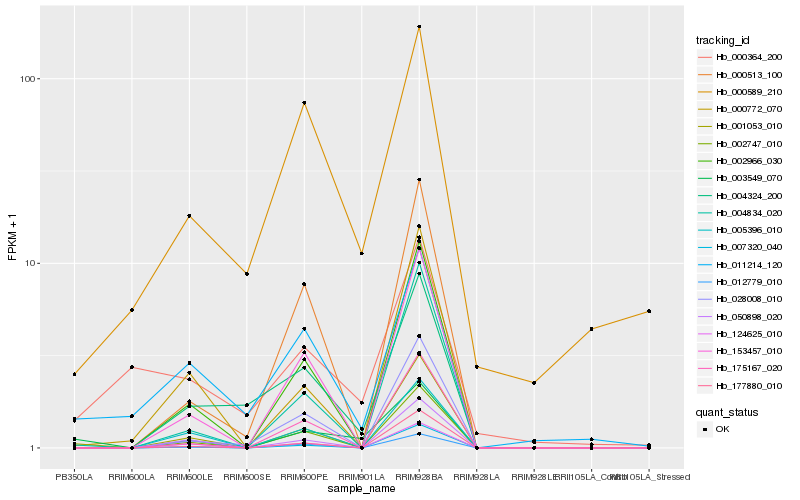
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003549_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_001053_010 |
0.2162748142 |
- |
- |
PREDICTED: polygalacturonase-like [Jatropha curcas] |
| 3 |
Hb_011214_120 |
0.2549681896 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 4 |
Hb_002747_010 |
0.2618279245 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 1 [Jatropha curcas] |
| 5 |
Hb_002966_030 |
0.2619685308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004834_020 |
0.2647519361 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 7 |
Hb_153457_010 |
0.2654547126 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 8 |
Hb_124625_010 |
0.2689008047 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 9 |
Hb_012779_010 |
0.2689314247 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 10 |
Hb_005396_010 |
0.2719163905 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 11 |
Hb_050898_020 |
0.2722859562 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 12 |
Hb_007320_040 |
0.2742810987 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 13 |
Hb_004324_200 |
0.2747814208 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 14 |
Hb_175167_020 |
0.2753368794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000589_210 |
0.2758008757 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 16 |
Hb_000364_200 |
0.2767388839 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000772_070 |
0.2788099448 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 18 |
Hb_000513_100 |
0.2798677611 |
- |
- |
germin-like protein GLP, partial [Manihot esculenta] |
| 19 |
Hb_028008_010 |
0.2801089406 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 20 |
Hb_177880_010 |
0.2803652011 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |