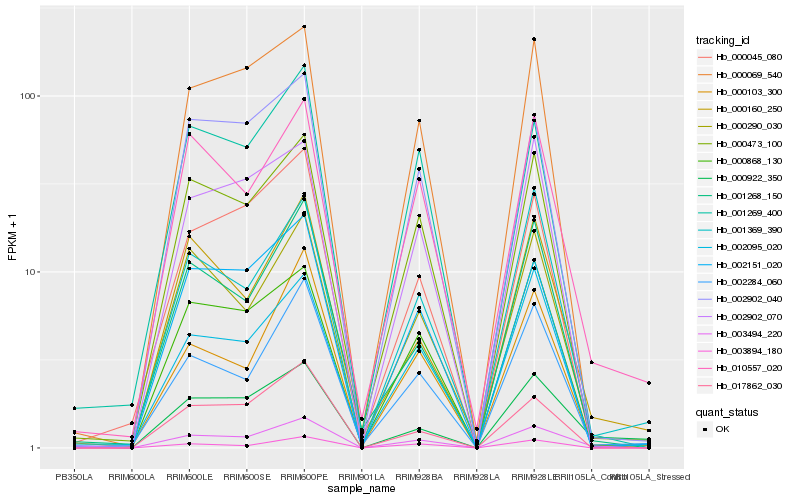
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003494_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 2 |
Hb_000473_100 |
0.1216694123 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_010557_020 |
0.1217954403 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 4 |
Hb_002284_060 |
0.1235238884 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_000290_030 |
0.1269394699 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 6 |
Hb_001268_150 |
0.1295764645 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000069_540 |
0.1303936044 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 8 |
Hb_000868_130 |
0.1304953068 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000160_250 |
0.1312816341 |
- |
- |
Uncharacterized protein TCM_019235 [Theobroma cacao] |
| 10 |
Hb_003894_180 |
0.1327319319 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 11 |
Hb_002151_020 |
0.1339019419 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 12 |
Hb_000922_350 |
0.1346407618 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 13 |
Hb_002095_020 |
0.1358226918 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001269_400 |
0.1369965363 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 15 |
Hb_002902_040 |
0.1372662543 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 16 |
Hb_000045_080 |
0.1380908066 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_017862_030 |
0.1392451343 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 18 |
Hb_001369_390 |
0.1394572628 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 19 |
Hb_000103_300 |
0.1397097918 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 20 |
Hb_002902_070 |
0.1402800825 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |