| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
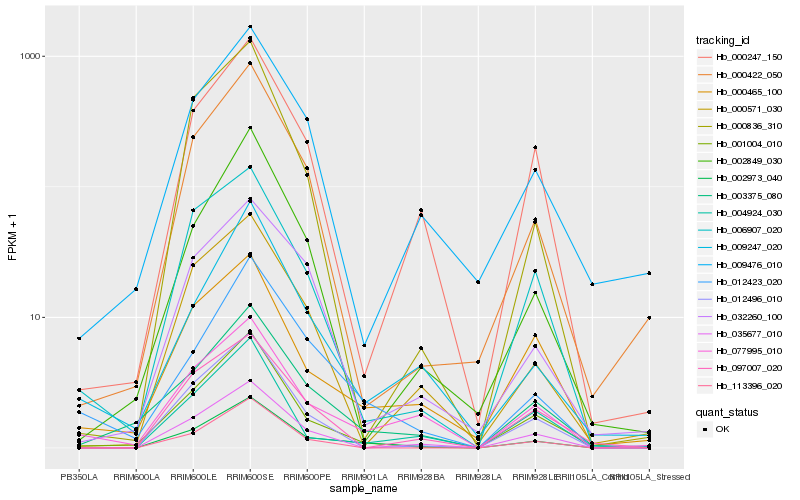
Hb_002973_040 |
0.0 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 2 |
Hb_001004_010 |
0.1427514927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003375_080 |
0.1498885993 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 4 |
Hb_035677_010 |
0.1520862705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012496_010 |
0.1546020749 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000247_150 |
0.1565014346 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 7 |
Hb_012423_020 |
0.1566787697 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 8 |
Hb_077995_010 |
0.1604613112 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_113396_020 |
0.168177114 |
- |
- |
Mitogen-activated protein kinase kinase kinase 1 [Glycine soja] |
| 10 |
Hb_000422_050 |
0.1683424266 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 11 |
Hb_000465_100 |
0.1693401203 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_004924_030 |
0.1711468701 |
- |
- |
- |
| 13 |
Hb_002849_030 |
0.1721007074 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 14 |
Hb_000571_030 |
0.1725237015 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 15 |
Hb_009247_020 |
0.1758397978 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 16 |
Hb_006907_020 |
0.1780109069 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase 3 [Manihot esculenta] |
| 17 |
Hb_097007_020 |
0.1808246781 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 18 |
Hb_032260_100 |
0.1811475309 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 19 |
Hb_000836_310 |
0.1812315408 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 30 [Jatropha curcas] |
| 20 |
Hb_009476_010 |
0.1822866628 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |